PCR-HRM分析篩查成骨不全一家系患兒基因突變
白雪李克秋任秀智何曉波王毅官士珍景亞青李光△
新技術(shù)交流
PCR-HRM分析篩查成骨不全一家系患兒基因突變
白雪1李克秋2任秀智3何曉波2王毅1官士珍1景亞青2李光2△
目的采用PCR-高分辨率熔解曲線(xiàn)(HRM)分析篩查成骨不全(OI)一家系患兒(先證者)COL1A1基因突變位點(diǎn),探討其基因型與臨床表型的聯(lián)系。方法對(duì)先證者進(jìn)行家族史及臨床資料的調(diào)查,采集先證者、家屬及50名正常對(duì)照者血液標(biāo)本,應(yīng)用PCR-HRM分析篩查先證者及正常對(duì)照者COL1A1基因突變,基因測(cè)序確證突變位點(diǎn)。結(jié)果先證者COL1A1基因17外顯子篩查結(jié)果異常,其熔解溫度(Tm)值比正常對(duì)照者Tm值低約0.4℃。先證者與正常對(duì)照者的標(biāo)準(zhǔn)熔解曲線(xiàn)及差異熔解曲線(xiàn)均有明顯差異。測(cè)序結(jié)果為c.1138G>A,突變導(dǎo)致380位氨基酸由甘氨酸(Gly)變成絲氨酸(Ser):p.(Gly 380 Ser),為錯(cuò)義突變。先證者父親、祖母均具有相同突變位點(diǎn)。先證者母親及正常對(duì)照者基因測(cè)序結(jié)果無(wú)此突變。該突變?cè)谥袊?guó)人群中未見(jiàn)報(bào)道。該家系遺傳特征為常染色體顯性遺傳,先證者臨床診斷為Ⅳ型OI,臨床表型較嚴(yán)重。結(jié)論P(yáng)CR-HRM分析是有效的OI基因篩查新方法。COL1A1基因c.1138G>A突變?cè)谥袊?guó)人群中為新發(fā)現(xiàn)的突變位點(diǎn)。α螺旋結(jié)構(gòu)域Gly被替換可能導(dǎo)致較嚴(yán)重的臨床表型。
成骨不全;遺傳篩查;聚合酶鏈反應(yīng);點(diǎn)突變;系譜;COL1A1基因;高分辨率熔解曲線(xiàn)分析
成骨不全(osteogenesis imperfecta,OI)又稱(chēng)脆骨病,是一種遺傳異質(zhì)性結(jié)締組織病,與Ⅰ型膠原缺陷相關(guān),患者因骨量減少、骨強(qiáng)度減弱,導(dǎo)致骨脆性增加,表現(xiàn)為易骨折、骨骼畸形及生長(zhǎng)缺陷。OI骨骼外表現(xiàn)包括:藍(lán)色鞏膜、牙質(zhì)形成不全和青春期后不同程度的聽(tīng)力損失等[1]。OI發(fā)病率約為1/10 000[2],90%以上OI患者由編碼Ⅰ型膠原α1/α2鏈的基因COL1A1/COL1A2突變引起[3]。OI的臨床表現(xiàn)輕重不等,從無(wú)明顯臨床癥狀到致死型均可發(fā)生[1]。基因檢測(cè)可為OI確診、產(chǎn)前診斷及遺傳咨詢(xún)提供依據(jù),但COL1A1/COL1A2基因突變?nèi)狈ν蛔儫狳c(diǎn),基因篩查工作量大、成本高。近年來(lái),高分辨率熔解曲線(xiàn)(high resolution melting,HRM)分析在醫(yī)學(xué)領(lǐng)域中發(fā)展迅速,已在多種疾病的基因突變檢測(cè)中證實(shí)其有效性[4-5]。本研究采用PCR-HRM分析篩查OI家系患兒(先證者)COL1A1基因突變位點(diǎn),探討其在OI基因研究中的作用。
1 資料與方法
1.1 一般資料收集1例2011年12月9日在我院住院的OI患兒(先證者)及其家系的臨床資料,包括家族史、骨折情況、骨骼畸形、非骨骼異常及X線(xiàn)檢查情況等。繪制家系圖譜,分析遺傳特征。OI家屬簽署“知情同意書(shū)”。先證者男,9歲,漢族。臨床診斷為Ⅳ型OI,其父母否認(rèn)近親婚配。先證者身高120 cm,體質(zhì)量23 kg,藍(lán)色鞏膜,牙質(zhì)形成不全,雞胸。患兒出生后38 d,即發(fā)生不明原因的上肢骨折,之后發(fā)生約30次四肢骨折,逐漸出現(xiàn)雙下肢向前向外彎曲畸形,自3歲后不能行走,雙下肢肌肉萎縮。X線(xiàn)片示:胸部畸形、左肱骨遠(yuǎn)段畸形、雙脛腓骨干可見(jiàn)彎曲畸形、雙股骨干畸形、關(guān)節(jié)畸形、骨干纖細(xì)、骨皮質(zhì)菲薄、骨質(zhì)密度低,胸部、上肢、雙下肢骨質(zhì)疏松,見(jiàn)圖1。家系調(diào)查顯示,先證者父親、祖母及家系中其他3位親屬均患有OI,見(jiàn)圖2。家系患者的臨床表型,見(jiàn)表1。
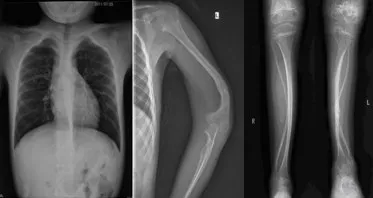
Fig.1 X-rays of the proband圖1 先證者X線(xiàn)片
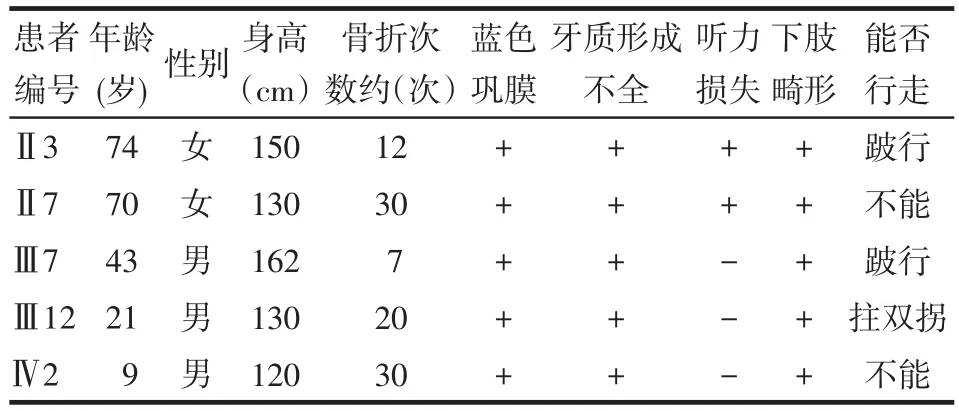
Tab.1 Clinical phenotype of OI family表1 OI家系患者的臨床表型
1.2 研究方法
1.2.1 基因組DNA制備采集先證者及家屬(先證者父親、母親、祖母)及50名正常對(duì)照者的靜脈血,EDTA抗凝。采用美國(guó)AxyPrep血基因組試劑盒,提取DNA標(biāo)本,使用Nano-Drop 2000核酸定量分析儀(德國(guó))檢測(cè)DNA濃度及純度,DNA標(biāo)本放置-80℃保存?zhèn)溆谩?/p>
1.2.2 引物設(shè)計(jì)及合成PCR-HRM引物設(shè)計(jì)參照Gentile 等[6]文獻(xiàn)報(bào)道,引物設(shè)計(jì)覆蓋了COL1A1基因的編碼區(qū)和相鄰的外顯子-內(nèi)含子交界處。擴(kuò)增產(chǎn)物大小平均149 bp,對(duì)較長(zhǎng)或有復(fù)雜熔解域的外顯子,選擇兩對(duì)或多對(duì)引物。引物由上海生工合成。
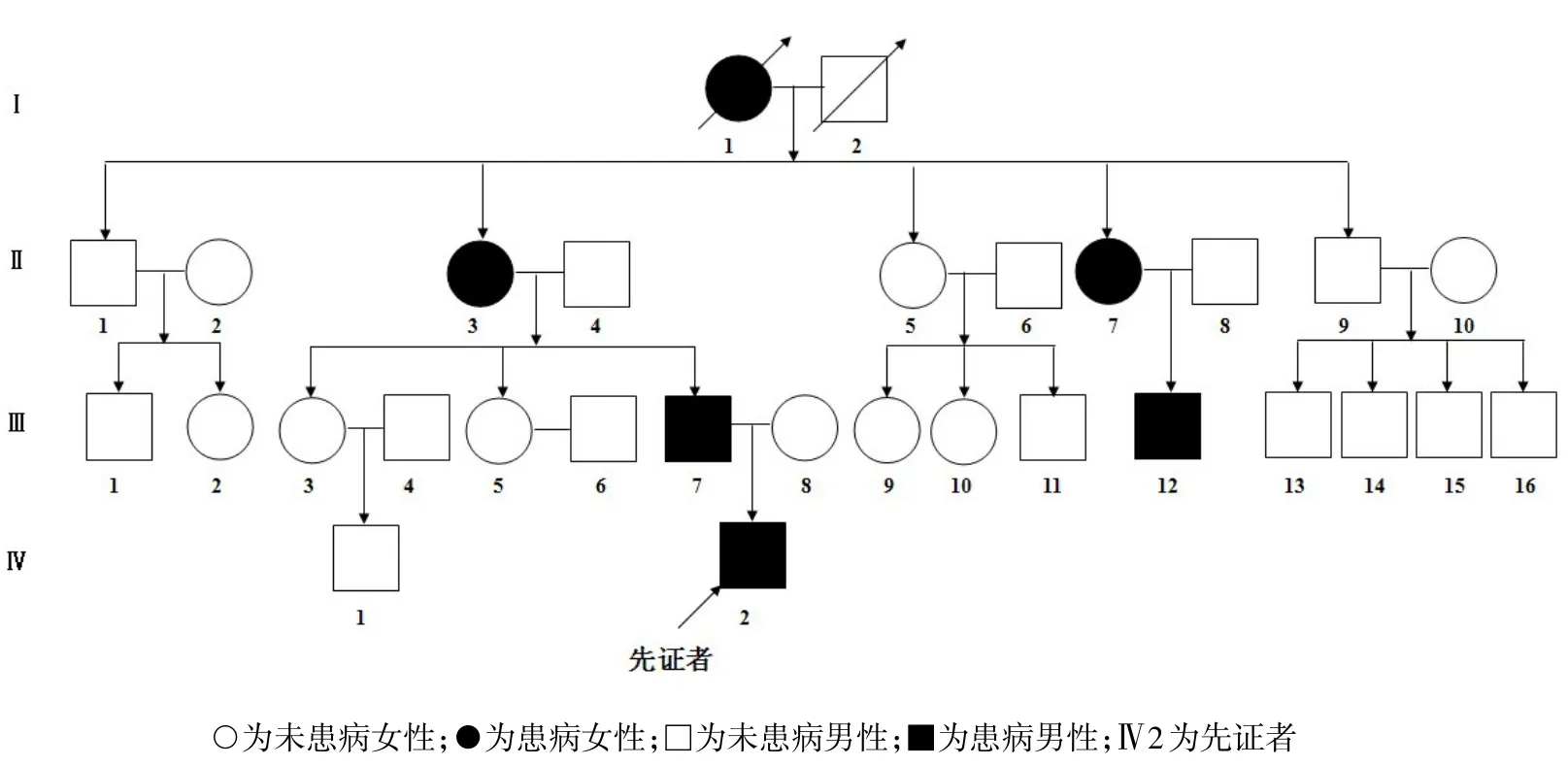
Fig.2 Pedigree chart圖2 家系圖
1.2.3PCR-HRM分析篩查COL1A1基因突變采用QIAGEN公司(德國(guó))Type-it HRM PCR Kit試劑盒,飽和染料為EvaGreen。采用基因公司(美國(guó))illumina Eco熒光定量PCR儀,反應(yīng)條件為:95℃預(yù)變性5 min;95℃變性10 s、退火溫度(58~64℃)30s、72℃延伸10s,40個(gè)循環(huán)。每份樣本總反應(yīng)體積為10 μL,PCRMasterMix 5μL,上、下游引物各10μmol/L0.7 μL;模板DNA及無(wú)酶水體積之和為3.6 μL,反應(yīng)體系模板DNA均為30 ng。HRM分析條件為95℃預(yù)處理15s,熔解溫度為65~95℃,溫度上升速率0.1℃/s,每升高1℃采集10次熒光信號(hào)數(shù)據(jù),Eco V3.0軟件對(duì)熔解曲線(xiàn)進(jìn)行分析,每個(gè)樣本重復(fù)2次。
1.2.4 長(zhǎng)片段引物的擴(kuò)增及測(cè)序針對(duì)PCR-HRM分析異常的基因區(qū)域自行設(shè)計(jì)長(zhǎng)片段引物,PCR-HRM分析先證者COL1A1基因17外顯子區(qū)域結(jié)果異常,設(shè)計(jì)長(zhǎng)片段引物,上游:5′-CAGCCCGTTCACTAACACCT-3′;下游:5′-ATTGGCACCTTTAGCACCAG-3′。采用Applied Biosystems公司梯度PCR儀(美國(guó)),測(cè)序由上海生工完成。
1.2.5 序列分析應(yīng)用BLAST及GENETOOL等生物信息學(xué)軟件分析測(cè)序結(jié)果,與GenBank上的參考序列進(jìn)行對(duì)比分析,確定突變位點(diǎn)及編碼氨基酸序列的變化。在人類(lèi)膠原突變數(shù)據(jù)庫(kù)(https://www.le.ac.uk/genetics/collagen/)中確定是否為新發(fā)現(xiàn)的突變位點(diǎn)。
2 結(jié)果
2.1PCR-HRM分析篩查COL1A1基因突變PCRHRM分析篩查顯示,先證者COL1A1基因17外顯子區(qū)域結(jié)果異常,先證者該區(qū)域擴(kuò)增片段的熔解溫度(Tm)值為88.6℃,正常對(duì)照平均Tm值為(89.0± 0.05)℃,兩者Tm值相差約0.4℃。標(biāo)準(zhǔn)熔解曲線(xiàn)提示,先證者COL1A1基因17外顯子區(qū)域存在雜合突變,見(jiàn)圖3。
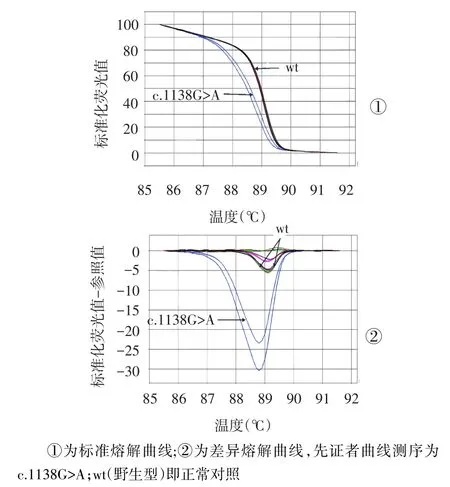
Fig.3 High resolution melting curve’s圖3 HRM熔解曲線(xiàn)
2.2 測(cè)序結(jié)果COL1A1基因17外顯子長(zhǎng)片段引物的擴(kuò)增產(chǎn)物為391 bp,先證者測(cè)序結(jié)果為:位于COL1A1基因17外顯子c.1138G>A雜合突變,也就是cDNA 1138位堿基G突變成A,先證者父親及祖母測(cè)序結(jié)果與先證者相同。先證者母親及正常對(duì)照測(cè)序結(jié)果與GenBank上參考序列相同,見(jiàn)圖4。
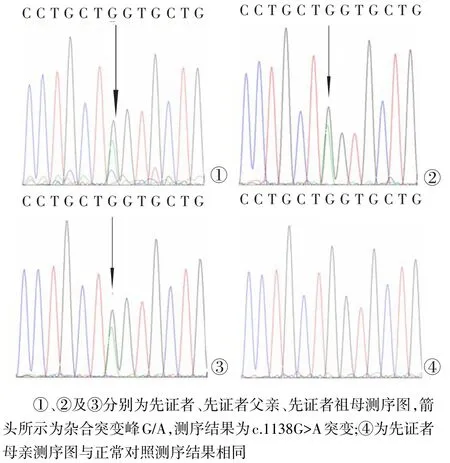
Fig.4 Sequencing curves of probands and their families圖4 先證者及家屬測(cè)序圖
3 討論
OI主要由COL1A1/COL1A2基因突變引起,該基因突變散布在整個(gè)基因,無(wú)突變熱點(diǎn)。目前已發(fā)現(xiàn)1 100多種COL1A1/COL1A2基因突變。突變形式有單個(gè)堿基置換、插入、缺失、重復(fù)、移碼和剪接位點(diǎn)等。通常每個(gè)家系有自己的“私有突變”[7]。Sanger測(cè)序被認(rèn)為是OI基因診斷的金標(biāo)準(zhǔn)[8],但測(cè)序所需工作量大、費(fèi)用高。2012年,Gentile等[6]首次采用定量PCR-HRM分析篩查OI患者COL1A1/COL1A2基因突變,表明該方法具有快速、成本低及靈敏度高等優(yōu)點(diǎn),適于OI基因測(cè)序前的突變篩查。HRM分析是新興的基因篩查和分型的遺傳學(xué)分析方法[9],該方法基于有序列變化的擴(kuò)增產(chǎn)物間微小的Tm值差異及NA片段熔解曲線(xiàn)的差異,進(jìn)行基因突變篩查,能夠區(qū)分單個(gè)堿基的變化[10]。本研究采用PCR-HRM方法篩查OI患兒COL1A1基因突變,優(yōu)化了PCRHRM分析條件,篩查出先證者COL1A1基因17外顯子Tm值較正常對(duì)照低約0.4℃。先證者標(biāo)準(zhǔn)熔解曲線(xiàn)的形狀與正常對(duì)照明顯不同,提示雜合突變。測(cè)序結(jié)果證實(shí),cDNA的1138位堿基為G/A的雜合突變。先證者父親及祖母均具有相同的突變位點(diǎn),先證者母親及正常對(duì)照該測(cè)序結(jié)果與GenBank上參考序列相同。結(jié)合該家系遺傳特征,先證者基因突變(c.1138G>A)來(lái)自其父親及祖母的遺傳。
目前,國(guó)內(nèi)對(duì)于OI的診斷主要根據(jù)臨床特征和影像學(xué)改變[11],仍以Sillence分型為主。Sillence等[12]根據(jù)患者臨床特征和組織病理學(xué)特性將OI分為Ⅰ~Ⅳ型,由COL1A1/COL1A2基因突變引起。Ⅰ型病情最輕,多無(wú)骨骼畸形,患兒身高正常或接近正常;Ⅱ型最嚴(yán)重,在母體子宮內(nèi)即發(fā)生骨折,常在圍產(chǎn)期死亡;Ⅲ型屬于非致死型中最嚴(yán)重的亞型,四肢進(jìn)行性畸形,大多患兒身材十分矮小,牙質(zhì)形成不全,脊椎壓縮和側(cè)彎。Ⅳ型病情屬于中度,臨床表現(xiàn)多與Ⅰ型和Ⅲ型相同,多數(shù)患者可下床行走,鞏膜著色,牙質(zhì)形成不全,聽(tīng)力受損和身材矮小的發(fā)生率變異較大[1]。骨折次數(shù)、骨骼畸形程度及牙質(zhì)形成不全通常與疾病的嚴(yán)重程度相關(guān)[3]。身材矮小或增長(zhǎng)不足是嚴(yán)重OI的又一關(guān)鍵特征[7]。本研究先證者及家系患者身高明顯低于正常人,骨折次數(shù)7~30次,均有藍(lán)色鞏膜、牙質(zhì)形成不全、下肢畸形,影響行走功能。先證者臨床診斷為Ⅳ型OI,家系中OI患者的整體臨床表型較為嚴(yán)重。
Ⅰ型膠原蛋白是骨骼、肌腱、韌帶和皮膚中的主要細(xì)胞外基質(zhì)蛋白。由2條α1鏈和1條α2鏈組成3股螺旋結(jié)構(gòu),分別由COL1A1和COL1A2基因編碼。每條鏈的螺旋區(qū)域含有338個(gè)連續(xù)重復(fù)的Gly(甘氨酸)-X-Y基本結(jié)構(gòu),Gly在該結(jié)構(gòu)中不可缺少,Gly側(cè)鏈殘基能調(diào)節(jié)膠原內(nèi)部3股螺旋的空間結(jié)構(gòu),對(duì)于螺旋正確的折疊至關(guān)重要。本研究先證者COL1A1基因17外顯子1138位的堿基G突變成A,使380位的Gly替換為Ser,該Gly位于α1鏈螺旋區(qū)域中。已有研究表明,Ⅰ型膠原α1和α2鏈三股螺旋結(jié)構(gòu)域中最常見(jiàn)的突變形式為Gly被Ser替代,使Ⅰ型膠原結(jié)構(gòu)異常,通常表現(xiàn)為臨床表型較為嚴(yán)重的Ⅱ~Ⅳ型[8,13-14]。另外,在分析基因型與臨床表型的聯(lián)系時(shí),還應(yīng)考慮環(huán)境因素及個(gè)體差異可能對(duì)表型的影響。目前,對(duì)于OI基因型和臨床表型間的具體關(guān)系還不清楚[13],相關(guān)研究及探討,將為其提供理論基礎(chǔ)。
[1]Forlino A,Cabral WA,Barnes AM,et al.New perspectives on osteogenesis imperfecta[J].Nat Rev Endocrinol,2011,7(9):540-557. doi:10.1038/nrendo.2011.81.
[2]Glorieux FH.Osteogenesis imperfecta[J].Best Pract Res Clin Rheumatol,2008,22(1):85-100.doi:10.1016/j.berh.2007.12.012.
[3]Bodian DL,Chan TF,Poon A,et al.Mutation and polymorphism spectrum in osteogenesis imperfecta type II:implications for genotype-phenotype relationships[J].Hum Mol Genet,2009 18(10): 1893-1895.doi:10.1093/hmg/ddn374.
[4]Yang J,Qian J,Lin J,et al.Development of a High-Resolution Melting Analysis for the Detection of the SF3B1 Mutations[J].Genet Test Mol Biomarkers,2013,17(4):342-347.doi:10.1089/gtmb.2012.0364.
[5]Pe?in I,Whittall R,Futema M,et al.Mutation detection in Croatian patients with familial hypercholesterolemia[J].Ann Hum Genet, 2013,77(1):22-30.doi:10.1111/j.1469-1809.2012.00735.x.
[6]Gentile FV,Zuntini M,Parra A,et al.Validation of a quantitative PCR-high-resolution melting protocol for simultaneous screening of COL1A1 and COL1A2 point mutations and large rearrangements: application for diagnosis of osteogenesis imperfect[J].Hum Mutat, 2012,33(12):1697-1707.doi:10.1002/humu.22146.
[7]Lee KS,Song HR,Cho TJ,et al.Mutational spectrum of type I collagen genes in Korean patients with osteogenesis imperfect[J].Hum Mutat,2006,27(6):599.
[8]van Dijk FS,Byers PH,Dalgleish R,et al.EMQN best practice guidelines for the laboratory diagnosis of osteogenesis imperfecta[J]. Eur J Hum Genet,2012,20(1):11-19.doi:10.1038/ejhg.2011.141.
[9]Rouleau E,Lefol C,Bourdon V,et al.Quantitative PCR high-resolution melting(qPCR-HRM)curve analysis,new approach to simultaneously screenpoint mutations and large rearrangements:application to MLH1 germline mutations in Lynch syndrome[J].Hum Mutat,2009,30(6):867-875.doi:10.1002/humu.20947.
[10]Skrzypczak-Zielinska M,Borun P,Milanowska K,et al.High-resolution melting analysis of the TPMT gene:a study in the Polish population[J].Genet Test Mol Biomarkers,2013,17(2):153-159.doi: 10.1089/gtmb.2012.0192.
[11]任秀智,曾裴,李冰,等.多段截骨矯形髓內(nèi)固定治療兒童成骨不全癥[J].中華骨科雜志,2012,32(5):477-481.
[12]Sillence DO,Senn A,Danks DM.Genetic heterogeneity in osteogenesis imperfect[J].J Med Genet,1979,16(2):101-116.
[13]Rauch F,Lalic L,Roughley P,et al.Relationship between genotype and skeletal phenotype in children and adolescents with osteogenesis imperfect[J].J Bone Miner Res,2010,25(6):1367-1374.doi: 10.1359/jbmr.091109.
[14]Marini JC,Forlino A,Cabral WA,et al.Consortium for osteogenesis imperfect mutations in the helical domain of type I collagen:regions rich in lethalmutations align with collagen binding sites for integrins and proteoglycans[J].Hum Mutat,2007,28(3):209-221.doi: 10.1359/jbmr.091109.
(2014-01-07收稿2014-02-21修回)
(本文編輯魏杰)
PCR-HRM Analysis for Gene Mutation Screening in a Child with Osteogenesis Imperfecta
BAI Xue1,LI Keqiu2,REN Xiuzhi3,HE Xiaobo2,WANG Yi1,GUAN Shizhen1,JING Yaqing2,LI Guang2
1 Department of Medical Laboratory,Tianjin Hospital,Tianjin 300211,China;2 Department of Biology,Tianjin Medical University;3 Department of Orthopedics Ward,Tianjin Wu Qing People′s Hospital
LI Guang,E-mail:lig@tijmu.edu.cn
ObjectiveTo investigate COL1A1 gene mutation by PCR-high resolution melting(PCR-HRM)and analyze the correlation between genotype and clinical phenotype in a child(proband)with osteogenesis imperfecta(OI).MethodsThe family history of OI pedigree along with the clinical data was collected.Blood samples from the proband and his family members,as well as 50 normal controls,were collected.The mutation of COL1A1 gene was screened using PCRHRM and validated by the gene sequence.ResultsThe detection of PCR-HRM showed the abnormal result of COL1A1 17 exon in proband with a lower melting temperature(Tm)value than that of normal controls by 0.4℃.There were significant differences in the standardization melting curve and the different melting curve between the proband and the normal controls.The sequencing result was c.1138G>A,which meant that cDNA of 1138 base G mutation into A.The mutations transformed the amino acid glycine into a serine at amino acid 380(P.Gly 380 Ser),which resulted in missense mutations.The proband’s father and grandmother had the same mutation of COL1A1 gene.The mutation was not found in the proband’s mother and normal controls.There was no report for such mutation in Chinese population.Pedigree analysis showed the family genetic characteristics of autosomal dominant inheritance.The proband was clinically diagnosed as OI typeⅣwith more severe clinical phenotype.ConclusionPCR-HRM analysis is a new effective method for genetic screening of OI.COL1A1 mutation of c.1138G>A is a newly discovered mutation in Chinese population.Gly replaced in α helical domain may lead to a more severe clinical phenotype.
osteogenesis imperfecta;genetic screening;polymerase chain reaction;point mutation;pedigree; COL1A1 gene;high-resolution melting
R394.112,R681.31
A
10.3969/j.issn.0253-9896.2014.07.020
國(guó)家高技術(shù)研究發(fā)展計(jì)劃(863計(jì)劃)資助項(xiàng)目(2012AA021003);國(guó)家自然科學(xué)基金資助項(xiàng)目(21177091);天津市衛(wèi)生局科技基金資助項(xiàng)目(2013KZ072)
1天津市天津醫(yī)院檢驗(yàn)科(郵編300211);2天津醫(yī)科大學(xué)生物學(xué)教研室;3天津市武清區(qū)人民醫(yī)院骨科三病區(qū)
△通訊作者E-mail:lig@tijmu.edu.cn

