廣西水稻地方品種核心種質稻瘟病抗性位點全基因組關聯分析
陳 燦 農保選 夏秀忠 張宗瓊 曾 宇 馮 銳 郭 輝 鄧國富 李丹婷 楊行海
廣西水稻地方品種核心種質稻瘟病抗性位點全基因組關聯分析
陳 燦**農保選**夏秀忠 張宗瓊 曾 宇 馮 銳 郭 輝 鄧國富 李丹婷*楊行海*
廣西農業科學院水稻研究所 / 廣西水稻遺傳育種重點實驗室, 廣西南寧 530007
稻瘟病是水稻重要病害之一, 嚴重影響水稻的產量與品質。培育抗性品種是防治稻瘟病最經濟、環保的方式。稻瘟病抗性基因的鑒定與挖掘是開展抗病育種的基礎與前提。本課題組前期對419份廣西水稻地方品種核心種質進行簡化基因組測序, 獲得208,993個高質量SNP標記。本研究采用苗期噴霧接種方法, 研究了該419份核心種質對7個稻瘟病生理小種的抗性, 并根據表型和基因型數據, 利用一般線性模型(general linear model, GLM)和混合線性模型(mixed linear model, MLM)進行全基因組關聯分析。2種模型下共檢測到20個位點, 其中GLM檢測到20個位點, MLM檢測到1個位點, Chr12_10803913位點在2種模型下都檢測到。17個位點與前人定位的基因/QTLs重疊, 其余3個是新位點, 分別為Chr3_18302718、Chr3_18302744及Chr5_10379127位點。在20個顯著關聯位點上下游各150 kb的基因組區域中共篩選出候選基因323個, 初步確定8個候選基因與抗病相關, 其中()、()為已知克隆的基因,、和為新位點附近篩選到的候選基因。本研究結果為稻瘟病抗性位點挖掘與稻瘟病相關基因克隆提供了科學依據。
水稻; 稻瘟病; 全基因組關聯分析; 候選基因
稻瘟病是由稻瘟菌()引起的一種水稻毀滅性真菌病害, 嚴重影響世界水稻種植區的產量與品質, 從而威脅全球糧食安全[1-2]。全球每年由稻瘟病引起的水稻產量損失約為總量的10%~30%, 它可以養活至少6000萬人, 經濟價值高達660億美元[3]。化學防治作為一種主要、傳統的病蟲害防治手段, 長期使用既污染環境, 也增加經濟成本。種質資源對抗性具有廣泛遺傳變異, 因此利用寄主植物自身抗性是防治該病最有效、最經濟和最環保的方法[4]。但病原真菌對宿主的適應能力的頻繁突變, 會使得品種抗性在3~5年內喪失[5]。因此, 需要不斷挖掘鑒定新的稻瘟病基因, 這對于開展抗病選育有重要的理論與實踐意義。
水稻抗性基因包括2種類型, 一種是提供種系特異性抗性的抗性基因(resistance gene, R), 另一種是控制部分非種系特異性抗性的微效基因, 又稱數量性狀位點(quantitative trait locus, QTL)。鑒定抗性基因(R)/數量性狀位點(QTL)及開展抗病機制的研究對抗病品種的選育尤為重要。水稻稻瘟病抗性基因挖掘一直是水稻抗病育種的熱點問題。迄今為止, 已經鑒定了110多個抗稻瘟病基因, 其中克隆了36個[6-8]。根據克隆基因的編碼蛋白類型, 可以分為5類。(1) 核苷酸結合位點(nucleotide binding site, NBS)-富含亮氨酸重復序列(leucine rich repeat, LRR)-蛋白(NBS-LRR或NLR), 根據N端結構域可分為2個子類TIR (Toll/Interleukin-1receptor)-NBS- LRR和CC (coiled-coil)-NBS-LRR。、、、、、、、、、、、、、、、等屬于前者,、、、、、、、、、、、、等屬于后者; (2) 凝集素受體(lectin receptor), 如; (3) 富含脯氨酸結構域蛋白(proline-rich domain proteins), 如; (4) 富含Armadillo重復序列蛋白, 如; (5) 富含四肽重復序列(tetratricopeptide repeats, TPRs)蛋白, 如。這些編碼蛋白類型為稻瘟病候選基因的篩選提供了重要理論依據。
大多數植物基因/QTL是通過連鎖作圖鑒定的, 因此基因/QTL的檢測受到使用的雙親材料的限制, 不能夠解釋自然界廣泛存在的遺傳變異。隨著高密度遺傳作圖技術的發展, 基于連鎖不平衡的全基因組關聯分析(genome-wide association analysis, GWAS)已成為自然群體基因/QTL鑒定的重要工具。GWAS克服了利用來自雙親材料的群體進行連鎖映射的缺點。通過與高通量測序、分群分析法(bulked segregant analysis, BSA)等技術手段相結合, GWAS在檢測水稻稻瘟病性狀關聯位點及篩選候選基因方面得到了大量快速應用[9-12]。如最近Li等[13]利用湖南省3個分離菌株和234份水稻多樣性小組1進行了水稻抗稻瘟病的全基因組關聯研究(GWAS), 共鑒定出56個QTL。其中1個QTL定位于抗性基因位點, 該位點對3個分離菌株均具有抗性。抗性品種基因組序列分析結果表明, 該位點是一個新的等位基因, 將其命名為。Lu等[14]利用全基因組關聯研究與RNA測序分析, 鑒定出127個水稻抗稻瘟病相關位點。此外, 在一個200 kb的基因組區域中預測了2341個非冗余候選基因, 其中45個基因與抗病相關。
本研究供試材料為419份廣西地方品種核心種質, 利用簡化基因組測序(specific-locus amplified fragment sequencing, SLAF-seq), 已經獲得了高質量的單核苷酸多態性位點(single nucleotide polymorphism, SNP) 208,993個[15]。利用該陣列SNP已對蒸煮食味、糯性、種皮顏色和南方黑條矮縮病等性狀進行了GWAS研究[15-18]。但利用GWAS檢測該群體的稻瘟病抗性位點尚未報道。
為此, 我們擬使用基于419份廣西地方品種核心種質的SNP數據, 并結合7個生理小種的苗期葉瘟抗性表型數據, 利用GWAS挖掘出該群體材料的稻瘟病關聯位點, 并預測顯著關聯位點附近區域的候選基因, 為下一步候選基因驗證及基因克隆提供理論依據。
1 材料與方法
1.1 供試材料及菌種
實驗材料來源于廣西農業科學院水稻研究所庫存的地方稻種資源核心種質, 共計419份, 包括330份秈稻、78份粳稻及11份其他類型品種。麗江新團黑谷和Tetep分別為感病及抗病對照。ZA 9、ZA 13、ZB 1、ZB 9、ZB 13、ZC 3和ZC 13等7個小種均由廣西農業科學院植物保護研究所提供。水稻材料及對照品種浸種催芽后播到塑料盤中, 每份材料播20粒, 待長至三至五葉齡時(大約2~3周)用配制好的孢子液人工噴霧接種。接種菌液濃度為1×105~2×105個 mL–1的孢子懸浮液[19], 然后在26℃及相對濕度95%左右的培養室培養24 h, 接種大約7 d后, 根據描述的病斑大小和面積比(diseased leaf area, DLA), 對其進行0到9分的評分[20]。每份材料鑒定3次。0~3級為抗病(resistance, 用R表示), 4~9級為感病(susceptibility, 用S表示), 取3次平均值為鑒定結果。利用SPSS 19統計分析軟件進行描述性統計及相關作圖分析。
1.2 SLAF測序和SNP基因分型
在Illumina Hiseq 2500系統上進行SLAF測序。利用BLAT軟件對clean reads進行聚類, 得到多態SLAF標簽。再使用BWA軟件, 將多態SLAF標簽序列比對至日本晴參考基因組上(http://plants. ensembl.org/Oryza_sativa/Info/Index)。然后利用GATK and SAM工具包分析SNP calling。根據最小等位基因頻率(minor allele frequency, MAF) > 0.05和完整性>0.5, 共獲得208,993個SNP[15]。
1.3 全基因組關聯分析的方法
使用軟件TASSEL V3對208,993個SNP基因型和苗期葉瘟表型數據進行GWAS關聯分析[21], 混合線性模型MLM為(Q+K)模型, Q為群體結構, K為親緣系數。采用MEGA5軟件構建系統發育樹。采用SPAGeDi軟件進行兩兩親屬關系分析。采用ADMIXTURE軟件分析種群結構[15]。< 4.79×10–6(1/208,993 = 4.79E-6)被認為具有顯著相關性。根據R環境生成Manhattan和Q-Q plot。
1.4 候選基因的選擇
以日本晴為參考基因組, 基于水稻的連鎖不平衡(linkage disequilibrium, LD)衰減情況, 參考楊行海等[16]研究結果, 選擇峰值SNP上下游各150 kb區間為候選基因區域。所有植物中的抗性基因, 包括NLR、凝集素受體等被認為是候選基因[22-23]。
2 結果與分析
2.1 苗期稻瘟病抗性評價
利用7個不同類型的稻瘟病生理小種對419份水稻材料進行苗期噴霧接種, 根據DLA評價其抗性, 并對抗性級別進行統計分析。DLA評估的抗性等級從“ZC13”的2.93到“ZA13”的5.72, 平均為4.49, 變異系數從“ZA13”的0.24到“ZC13”的0.48, 平均為0.34 (表1)。根據抗性級別均值, 可以發現ZA種群毒性最強, 其次是ZB種群, ZC種群較弱, 這與實際情況相一致, 這說明這3類小種對實驗材料有很好的鑒別性。從圖1可以看出, 在接種7個不同類型的稻瘟病生理小種下, 葉瘟抗性級別分布具有較好的擬合正態分布, 利于開展GWAS關聯分析。
2.2 全基因組關聯分析
親緣關系與群體結構分析表明, 系統發育樹聚集為2個主要類群, 其與秈稻、粳稻亞群相一致。419份地方種質分為6個類型群體結構, 包括秈稻、粳稻亞類[15]。
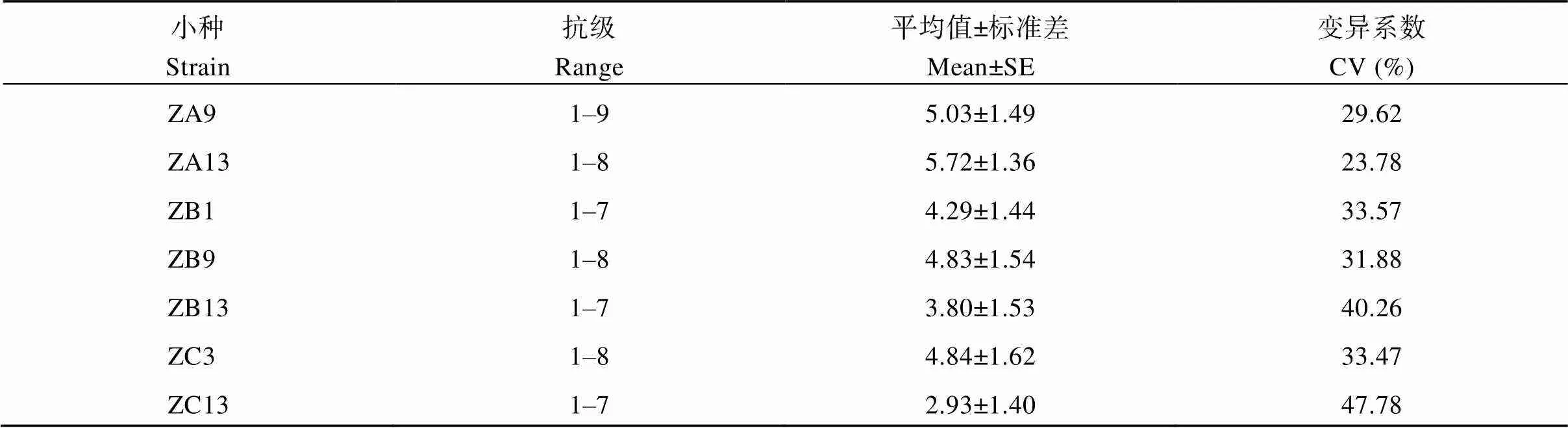
表1 不同稻瘟病小種接種下水稻苗期葉瘟抗性級別統計分析
在<4.79×10–6(4.79E-6)水平下, 使用一般線性模型(GLM)共檢測到20個稻瘟病相關SNP (圖2-A~C和表2), 其中位點10,803,913在MLM下也被檢測到, 3個位點為新位點, 分別為3號染色體上的18,302,718、18,302,744位點(Chr3_18302718, Chr3_18302744)及5號染色體上10,379,127位點(Chr5_10379127), 其他17個位點附近均有已知定位基因/QTL。在接種的7個小種中, 只有3個小種關聯到顯著性位點, 平均每個小種關聯到6.7個位點。ZC3關聯到最多的位點, 共16個, ZC13、ZB9各關聯到2個位點。從關聯位點在染色體上的分布來看, 12號染色體關聯的位點最多, 為13個, 其次是1號和3號染色體, 分別為3個和2個, 5號和8號染色體最少, 均為1個。
在<4.79×10–6(4.79E-6)水平下, 使用混合線性模型(MLM)僅檢測到1個稻瘟病相關SNP (圖2-D和表2), 該位點10,803,913位于12號染色體(Chr12_10803913)。
2.3 候選基因分析
根據水稻基因組注釋(http://rice.plantbiology. msu.edu/)及LD衰退水平, 在20個關聯位點300 kb的基因組區域中共鑒定出候選基因323個。根據已知克隆稻瘟病抗性基因的編碼蛋白類型、候選基因功能注釋信息及利用植物比較基因組學資源庫(https://phytozome.Jgi.doe.gov/pz/portal.html)蛋白序列同源比對分析, 初步篩選到8個候選基因與稻瘟病相關, 其中()、()為已知克隆基因(表3)。本研究鑒定到的一些SNP位點與這2個克隆基因距離非常接近。如12號染色體上的位點10,629,609 (= 6.56E-08)與[24-25](10,606,359~10,612,068)相距僅約17.5 kb; 該染色體上的另一個位點10,816,338 (= 8.01E-09)與[26](10,822,534~10,833,768)相距僅約6.2 kb。和功能注釋為病理相關蛋白,編碼含NB-ARC結構蛋白。說明這3個基因可能是抗病候選基因。
A、B、C分別表示GLM模型ZB9、ZC3、ZC13的曼哈頓圖。D表示MLM模型ZC3的曼哈頓圖。E、F、G分別表示GLM模型ZB9、ZC3、ZC13的QQ圖。H表示MLM模型ZC3的QQ圖。圖中實心倒三角形為本文中顯著性關聯位點。
A, B, and C represent Manhattan plot of GLM model ZB9, ZC3, and ZC13, respectively. D represents Manhattan plot ZC3 in MLM model. D, E, and F represent Quantitle-Quantitle plot of GLM model ZB9, ZC3, and ZC13, respectively. H represents Quantitle-Quantitle plot ZC3 in MLM model. The solid inverse triangle in the figure is the significant correlation point in this study.
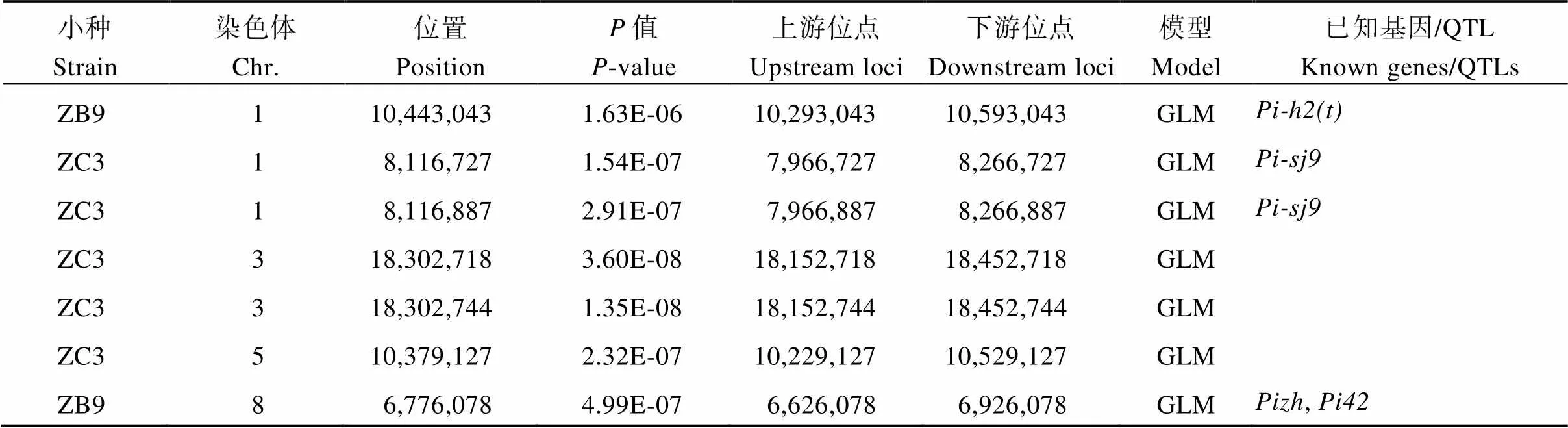
表2 水稻稻瘟病顯著關聯的SNP位點及已定位的基因/QTL
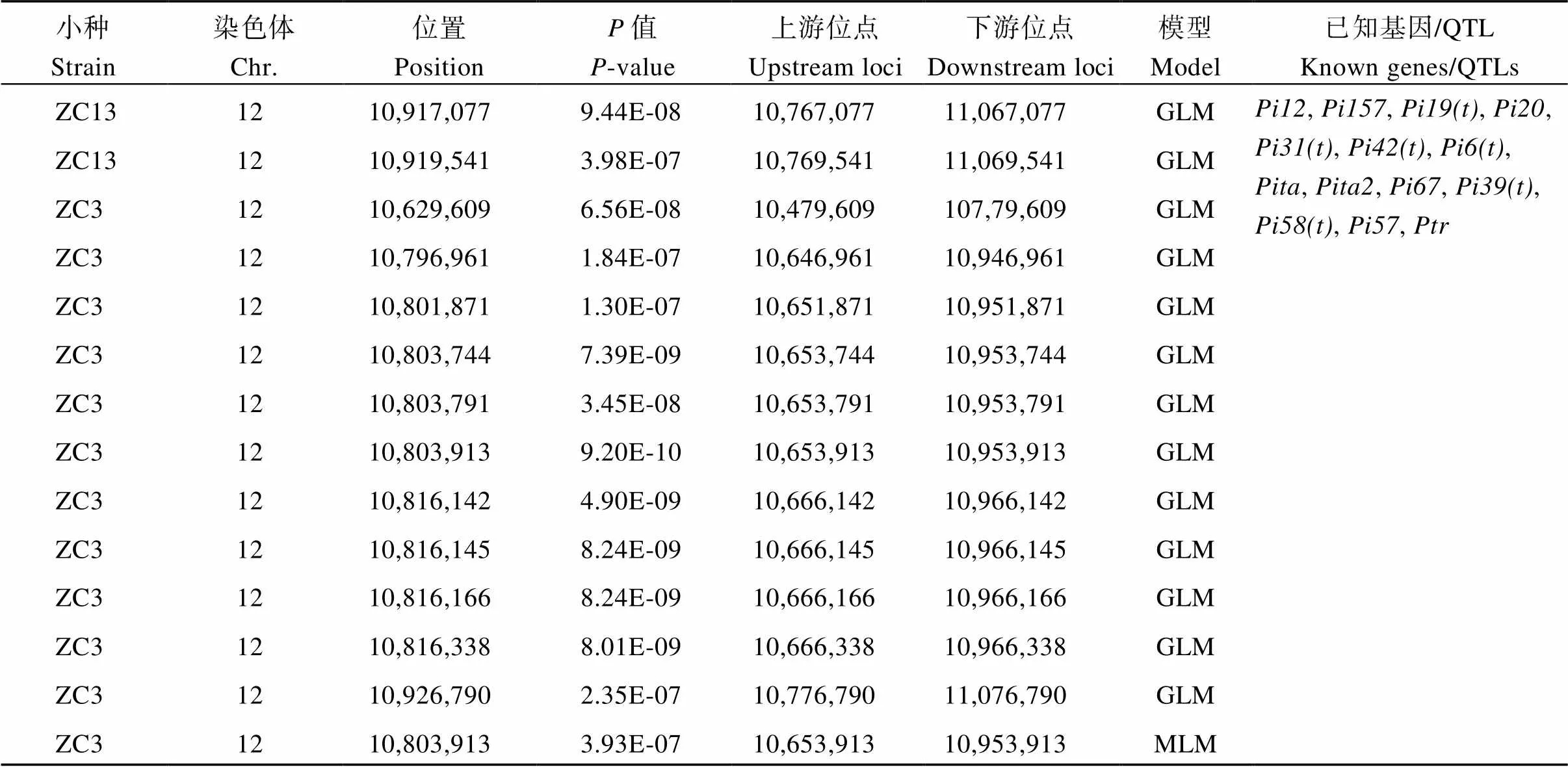
(續表2)
通過蛋白序列同源比對, 在3個新的關聯位點附近篩選到3個可能與稻瘟病相關的候選基因, 分別為、和。編碼蛋白與玉米的2個基因編碼產物相似度為66.8%、69.0%, 這2個基因都與ARM重復超家族蛋白相關, 推測此基因可能屬于含ARM重復序列蛋白抗病類型。編碼蛋白與菠蘿的1個基因編碼產物相似度為60.8%。該基因編碼富亮氨酸重復受體蛋白激酶家族蛋白, 推測此基因可能屬于受體蛋白激酶抗病類型。編碼蛋白與玉米的1個基因編碼產物相似度為57.9%。該基因編碼富亮氨酸重復跨膜蛋白激酶, 推測此基因可能屬于受體蛋白激酶抗病類型。
3 討論
GWAS已廣泛應用于植物各種性狀的基因位點挖掘。在稻瘟病抗性基因方面, 前人已經開展了大量的研究工作, 并關聯到一些與已知基因/QTL相重疊的顯著性關聯位點[9-14]。這些基因/QTL主要是、、和等。本研究也在這些基因/QTL附近關聯到一些顯著性位點, 這說明這些基因/QTL可能廣泛存在于自然群體材料中。
在1號染色體上, 本研究共鑒定出3個顯著關聯位點, 這些位點與該染色體上的2個基因相重疊(圖3-A), 分別是[27](7.86 Mb~4.97 Mb)和[28](8.41 Mb~18.83 Mb)。在8號染色體上, 本研究共鑒定出1個顯著關聯位點, 該位點與該染色體上的2個基因相重疊, 分別是[29](4.37 Mb~21.01 Mb)和[30](5.11 Mb~6.76 Mb)。在12號染色體上, 本研究共鑒定出13個顯著關聯位點, 這些位點與該染色體上的14個基因相重疊或位于其附近(圖3-B)其分別為:[24-25](10.60 Mb~10.60 Mb)、[26](10.82 Mb~10.83 Mb)、[31](7.73 Mb~11.91 Mb)、[32](6.98 Mb~15.12 Mb)、[33](8.82 Mb~18.05 Mb)、[34](10.73 Mb~12.04 Mb)、[35](6.98 Mb~10.60 Mb)、[36](9.36 Mb~12.24 Mb)、[37](4.05 Mb~18.86 Mb)、[38](10.07 Mb~13.21 Mb)、[39](10.60 Mb~ 12.63 Mb)、[40](10.61 Mb~10.65 Mb)、[41](7.46 Mb~11.26 Mb)和[42](10.79 Mb~10.85 Mb)。以上說明本研究群體可能含有上述基因。
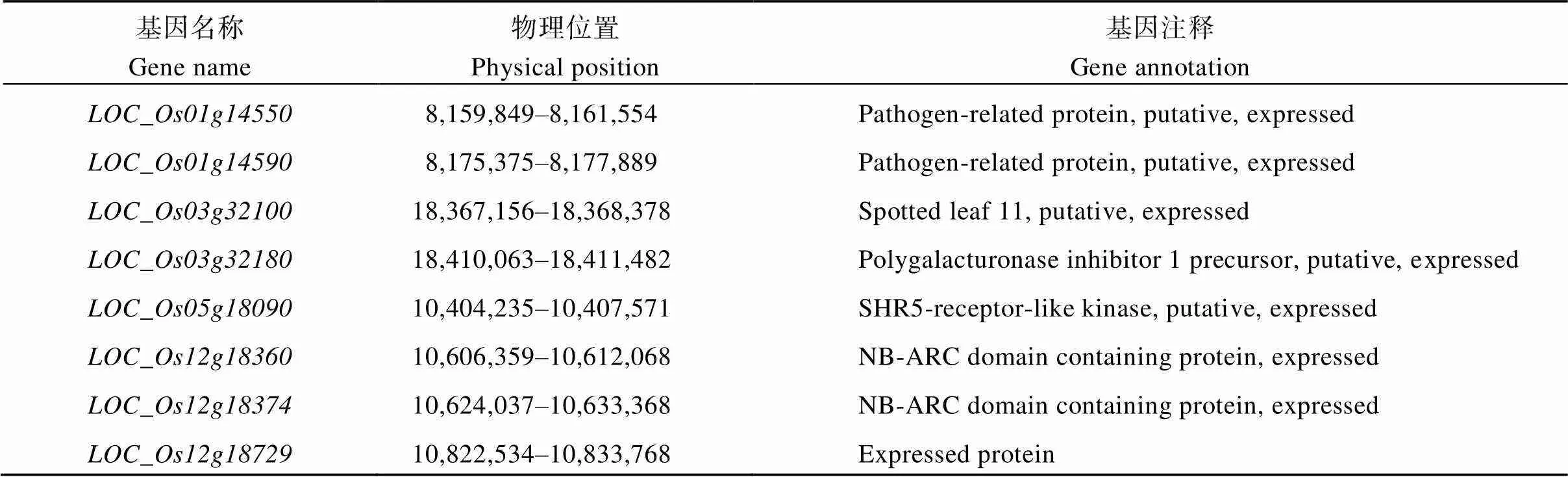
表3 候選基因信息
在3號染色體及5號染色體上, 本研究分別鑒定出2個(Chr3_18302718、Chr3_18302744)和1個(Chr5_ 10379127)顯著關聯位點, 這些位點都與染色體上已經定位的基因([43](22.92 Mb~27.89 Mb)和[44](9.33 Mb~9.78 Mb)在3號染色體上;[45](14.52 Mb~18.85 Mb)、[46](10.75 Mb~19.17 Mb)和[31](2.06 Mb~2.76 Mb)在5號染色體上)不重疊。據此推測, 這些位點附近可能含有新的抗稻瘟病基因。
進一步候選基因分析表明, 初步篩選到了8個與稻瘟病相關的候選基因, 其中()()為已知克隆基因。也被其他研究者認為是稻瘟病相關的候選基因[47], 這與本研究結果相一致。和為新關聯位點附近篩選到的候選基因, 其中在接種稻瘟菌后基因表達量顯著上調[48]。其他候選基因如和在接種稻瘟菌后基因表達量也顯著上調[48-49], 其中()是一個非小種特異性的C2H2轉錄因子, 協調減弱過氧化氫的降解, 表現出對稻瘟病的廣譜抗性[50]。本研究發現, 雖然在關聯位點Chr3_18302718和Chr3_18302744的區間內, 但是該位點僅在接種ZC3小種后鑒定到, 即該位點上的候選基因不具有廣譜抗性, 故不含有。以上候選基因是否在本研究中參與稻瘟病抗性過程,還需要作進一步的實時定量PCR驗證。
倒三角形所指關聯位點或關聯位點所在區域。A: 1號染色體上已定位的稻瘟病基因; B: 12號染色體上與顯著關聯位點區域重疊的稻瘟病基因。
An inverted triangle refers to the region of an association point or associated bit. A: the rice blast genes located on chromosome 1; B: the rice blast genes on chromosome 12 overlapped with the significantly associated locus region.
4 結論
基于GLM和MLM模型的GWAS結果, 共檢測到20個與稻瘟病抗性顯著相關的SNP位點, 其中17個位點與前人定位的基因/QTLs重疊, 其余3個是新位點。在關聯位點區域初步確定8個候選基因與抗病相關, 包括2個克隆的基因()、()及3個新位點附近篩選到的基因、和。
[1] Ashkani S, Rafii M Y, Shabanimofrad M, Miah G, Sahebi M, Azizi P, Tanweer F A, Akhtar M S, Nasehi A. Molecular breeding strategy and challenges towards the improvement of blast disease resistance in rice crop., 2015, 6: 886.
[2] Sakulkoo W, Osés-Ruiz M, Garcia E O, Soanes D M, Littlejohn G R, Hacker C, Correia A, Valent B, Talbot N J. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus., 2018, 359: 1399–1403.
[3] Scheuermann K K, Raimondi J V, Marschalek R, Andrade A D, Wickert E. The Molecular Basis of Plant Genetic Diversity. Shanghai: InTech China, 2012. pp 331–356.
[4] Manandhar H K, Lyngs Jorgensen H J, Mathur S B, Smedegaard-Peterson V. Suppression of rice blast by preinoculation with avirulentand the nonrice pathogen., 1998, 88: 735–739.
[5] Zeigler R S, Leong S A, Teng P S. Rice Blast Disease. Wallingford: CAB International, 1994. pp 626.
[6] 曹妮, 陳淵, 季芝娟, 曾宇翔, 楊長登, 梁燕. 水稻抗稻瘟病分子機制研究進展. 中國水稻科學, 2019, 33: 489–498. Cao N, Chen Y, Ji Z J, Zeng Y X, Yang C D, Liang Y. Recent progress in molecular mechanism of rice blast resistance., 2019, 33: 489–498 (in Chinese with English abstract).
[7] Zheng C Q, Jiang N, Zhao X H, Yan T Z, Fu J, Li Y F, Wu Z X, Hu X C, Bai Z N, Liu T G, Xiao G, Zhou Y B, Chen L B, Wang K, Yang Y Z. Identification of the blast resistance genefrom Chaling common wild rice (Griff)., 2020, 168: 211–219.
[8] Kalia S, Rathour R. Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice., 2019, 9: 209.
[9] Mgonja E M, Park C H, Kang H X, Balimponya E G, Opiyo S, Bellizzi M, Mutiga S K, Rotich F, Ganeshan V D, Mabagala R, Sneller C, Correll J, Zhou B, Talbot N J, Mitchell T K, Wang G L. Genotyping-by-sequencing-based genetic analysis of African rice cultivars and association mapping of blast resistance genes againstpopulations in Africa., 2017, 107: 1039–1046.
[10] Wang C H, Yang Y L, Yuan X P, Xu Q, Feng Y, Yu H Y, Wang Y P, Wei X H. Genome-wide association study of blast resistance inrice., 2014, 14: 311–321.
[11] Kang H X, Wang Y, Peng S S, Zhang Y L, Xiao Y H, Wan D, Qu S H, Li Z Q, Yan S Y, Wang Z L, Liu W D, Ning Y S, Korniliev P, Leung H, Mezey J, Mccouch S R, Wang G L. Dissection of the genetic architecture of rice resistance to the blast fungus., 2016, 17: 959–972.
[12] Lin H A, Chen S Y, Chang F Y, Tung C W, Chen Y C, Shen W C, Chen R S, Wu C W, Chung C L. Genome-wide association study of rice genes and loci conferring resistance toisolates from Taiwan., 2018, 59: 32.
[13] Li C G, Wang D, Peng S S, Chen Y, Su P, Chen J B, Zheng L M, Tan X Q, Liu J L, Xiao Y H, Kang H X, Zhang D Y, Wang G L, Liu Y. Genome-wide association mapping of resistance against rice blast strains in South China and identification of a newallele., 2019, 12: 47.
[14] Lu Q, Wang C H, Niu X J, Zhang M C, Xu Q, Feng Y, Yang Y L, Wang S, Yuan X P, Yu H Y, Wang Y P, Wei X H. Detecting novel loci underlying rice blast resistance by integrating a genome-wide association study and RNA sequencing., 2019, 39: 81.
[15] Yang X H, Xia X Z, Zeng Y, Nong B X, Zhang Z Q, Wu Y Y, Xiong F Q, Zhang Y X, Liang H F, Deng G F, Li D T. Identification of candidate genes for gelatinization temperature, gel consistency and pericarp color by GWAS in rice based on SLAF-sequencing., 2018, 13: e0196690.
[16] 楊行海, 農保選, 夏秀忠, 張宗瓊, 曾宇, 劉開強, 鄧國富, 李丹婷. 水稻糯性相關基因的全基因組關聯分析. 植物學報, 2016, 51: 737–742. Yang X H, Nong B X, Xia X Z, Zhang Z Q, Zeng Y, Liu K Q, Deng G F, Li D T. Genome-wide association study of genes related to waxiness in., 2016, 51: 737–742 (in Chinese with English abstract).
[17] 楊行海, 農保選, 夏秀忠, 張宗瓊, 曾宇, 劉開強, 鄧國富, 李丹婷. 廣西地方稻種資源核心種質紅色種皮全基因組關聯分析及鑒定兩個新的等位基因. 分子植物育種, 2017, 15: 1–6. Yang X H, Nong B X, Xia X Z, Zhang Z Q, Zeng Y, Liu K Q, Deng G F, Li D T. Validation of the red pericarp gene from 419 rice landrace core collection in Guangxi using genome-wide association study and discovery of two novelalleles., 2017, 15: 1–6 (in Chinese with English abstract).
[18] 農保選, 秦碧霞, 夏秀忠, 楊行海, 張宗瓊, 曾宇, 劉馳, 蔡健和, 謝慧婷, 崔麗賢, 羅群昌, 鄧國富, 劉丕慶, 李丹婷. 南方水稻黑條矮縮病苗期抗性的全基因組關聯分析. 分子植物育種, 2019, 17: 1069–1079. Nong B X, Qin B X, Xia X Z, Yang X H, Zhang Z Q, Zeng Y, Liu C, Cai J H, Xie H T, Cui L X, Luo Q C, Deng G F, Liu P Q, Li D T. Genome-wide association study of seedling resistance ofstreaked dwarf virus., 2019, 17: 1069–1079 (in Chinese with English abstract).
[19] Park C H, Chen S B, Shirsekar G, Zhou B, Khang C H, Songkumarn P, Afzal A J, Ning Y S, Wang R Y, Bellizzi M, Valent B, Wang G L. Theeffector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice., 2012, 24: 4748–4762.
[20] IRRI (International Rice Research Institute). Standard Evaluation System for Rice. Philippines: International Rice Research Institute, Manila, Philippines. 1996. pp 17–18.
[21] Zhang Z W, Ersoz E, Lai C Q, Todhunter R J, Tiwari H K, Gore M A, Bradbury P J, Yu J M, Arnett D K, Ordovas J M, Buckler E S. Mixed linear model approach adapted for genome-wide association studies., 2010, 42: 355–360.
[22] Liu W D, Liu J L, Triplett L, Leach J E, Wang G L. Novel insights into rice innate immunity against bacterial and fungal pathogens., 2014, 52: 213–241.
[23] Kourelis J, van der Hoorn R A L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function., 2018, 30: 285–299.
[24] Orbach M J, Farrall L, Sweigard J A, Chumley F G, Valent B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene., 2000, 12: 2019–2032.
[25] Bryan G T, Wu K S, Farrall L, Jia Y L, Hershey H P, McAdams S A, Faulk K N, Donaldson G K, Tarchini R, Valent B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene., 2000, 12: 2033–2045.
[26] Zhao H J, Wang X Y, Jia Y L, Minkenberg B, Wheatley M, Fan J B, Jia M H, Famoso A, Edward J D, Wamishe Y, Valent B, Wang G L, Yang Y N. The rice blast resistance geneencodes an atypical protein required for broad-spectrum disease resistance., 2018, 9: 2039.
[27] 宋微. 松粳9號對稻瘟病抗性及抗病基因定位. 東北農業大學碩士學位論文, 黑龍江哈爾濱, 2013. Song W. Identification and Gene Mapping of Resistance toin Songjing No. 9. MS Thesis of Northeast Agricultural University, Harbin, Heilongjiang, China, 2013 (in Chinese with English abstract).
[28] Xiao W M, Yang Q Y, Sun D Y, Wang H, Guo T, Liu Y Z, Zhu X Y, Chen Z Q. Identification of three majorgenes responsible for broad-spectrum blast resistance in anrice accession., 2015, 35: 49.
[29] Causse M A, Fulton T M, Cho Y G, Ahn S N, Chunwongse J, Wu K S, Xiao J H, Yu Z H, Ronald P C, Harrington S E, Second G, McCouch S R, Tanksley S D. Saturated molecular map of the rice genome based on an interspecific backcross population., 1994, 138: 1251–1274.
[30] Lee S, Wamishe Y, Jia Y, Liu G, Jia M H. Identification of two major resistance genes against race IE-1k oftherice cultivar Zhe 733., 2009, 24: 127–134.
[31] Sallaud C, Lorieux M, Roumen E, Tharreau D, Berruyer R, Svestasrani P, Garsmeur O, Ghesquiere A, Notteghem J L. Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy., 2003, 106: 794–803.
[32] Zheng K L, Zhuang J Y, Lu J, Qian H R, Lin H X. Identification of DNA markers tightly linked to blast resistance genes in rice. In: Khush G S, Hettel G, Rola T, eds. Rice Genetics III (in Part 2), IRRI, Manila, Philippines, 2008. pp 565–569.
[33] Naqvi N I, Chattoo B B. Molecular genetic analysis and sequence characterized amplified region-assisted selection of blast resistance in rice., 1996, 3: 570–576.
[34] Koide Y, Telebanco-Yanoria M J, Pena F D, Fukuta Y, Kobayashi N. Characterization of rice blast isolates by the differential system and their application for mapping a resistance gene,., 2011, 159: 85–93.
[35] Li W, Lei C L, Cheng Z J, Jia Y L, Huang D L, Wang J L, Wang J K, Zhang X, Su N, Guo X P, Zhai H Q, Wan J M. Identification of SSR markers for a broad-spectrum blast resistance genefor marker-assisted breeding., 2008, 22: 141–149.
[36] Kumar P, Pathania S, Katoch P, Sharma T R, Plaha P, Rathour R. Genetic and physical mapping of blast resistance geneon the short arm of rice chromosome 12., 2010, 25: 217–228.
[37] Yu Z H, Mackill D J, Bonman J M, McCouch S R, Guiderdoni E, Notteghem J L, Tanksley S D. Molecular mapping of genes for resistance to rice blast (Sacc.)., 1996, 93: 859–863.
[38] Hayashi K, Yoshida H, Ashikawa I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes., 2006, 113: 251–260.
[39] Joshi S, Dhatwalia S, Kaachra A, Sharma K D, Rathour R. Genetic and physical mapping of a new rice blast resistance specificityfrom a broad spectrum resistant genotype Tetep., 2019, 215: 9.
[40] Liu X Q, Yang Q Z, Lin F, Hua L X, Wang C T, Wang L, Pan Q H. Identification and fine mapping of, a major gene conferring the broad-spectrum resistance to., 2007, 278: 403–410.
[41] Koide Y, Telebanco-Yanoria M J, Fukuta Y, Kobayashi N. Detection of novel blast resistance genes,and, in a Myanmar rice landrace based on a standard differential system., 2013, 32: 241–252.
[42] Dong L Y, Liu S F, Xu P, Deng W, Li X D, Tharreau D, Li J, Zhou J W, Wang Q, Tao D Y, Yang Q Z. Fine mapping of() conferring broad spectrum resistance againstin introgressionline IL-E1454 derived from., 2017, 12: e0186201.
[43] Liang Z J, Wang L, Pan Q H. A new recessive gene conferring resistance against rice blast., 2016, 9: 47.
[44] Devi S J S R, Singh K, Umakanth B, Vishalakshi B, Rao K V S, Suneel B, Sharma S K, Kadambari G K M, Prasad M S, Senguttvel P, Syamaladevi D P, Madhav M S. Identification and characterization of a large effect QTL fromrevealedas putative candidate gene for rice blast resistance., 2020, 13: 17.
[45] Naqvi N I, Bonman J M, Mackill D J, Nelson R J, Chattoo B B. Identifcation of RAPD markers linked to a major blast resistance gene in rice., 1995, 1: 341–348.
[46] Ahn S N, Kim Y K, Hong H C, Han S S, Choi H C, McCouch S R, Moon H P. Mapping of genes conferring resistance to Korean isolates of rice blast fungus using DNA markers.. 1997, 29: 416–423.
[47] 魯清. 水稻種質資源重要農藝性狀的全基因組關聯分析. 中國農業科學院博士學位論文, 北京, 2016. Lu Q. Genome-wide Association Studies of Important Agronomic Traits in Rice Germplasm. PhD Dissertation of Chinese Academy of Agricultural Sciences, Beijing, China, 2016 (in Chinese with English abstract).
[48] Bagnaresi P, Biselli C, Orrù L, Urso S, Crispino L, Abbruscato P, Piffanelli P, Lupotto E, Cattivelli L, Vale G. Comparative transcriptome profiling of the early response toin durable resistant vs susceptible rice (L.) genotypes., 2012, 7: e51609.
[49] Meng Q F, Gupta R, Kwon S J, Wang Y M. Agrawal G K, Rakwal R, Park S R, Kim S T. Transcriptomic analysis ofleaves reveals key changes in response toMSP1., 2018, 34: 257.
[50] Li W T, Zhu Z W, Chern M, Yin J J, Yang C, Ran L, Cheng M P, He M, Wang K, Wang J, Zhou X G, Zhu X B, Chen Z X, Wang J C, Zhao W, Ma B T, Qin P, Chen W L, Wang Y P, Liu J L, Wang W M, Wu X J, Li P, Wang J R, Zhu L H, Li S G, Chen X W. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance., 2017, 170: 114–126.
Genome-wide association study of blast resistance loci in the core germplasm of rice landraces from Guangxi
CHEN Can**, NONG Bao-Xuan**, XIA Xiu-Zhong, ZHANG Zong-Qiong, ZENG Yu, FENG Rui, GUO Hui, DENG Guo-Fu, LI Dan-Ting*, and YANG Xing-Hai*
Rice Research Institute, Guangxi Academy of Agricultural Sciences / Guangxi Key Laboratory of Rice Genetics and Breeding, Nanning 530007, Guangxi, China
Blast disease is one of the most important rice diseases, which seriously affects the yield and quality in rice. In general, breeding resistant varieties is the most economical, environmental, and friendly way to control rice blast. Identification and mining of blast resistance genes are the basis and premise of disease resistance breeding. In our previous study, 419 core germplasms from Guangxi rice landraces were sequenced using specific-locus amplified fragment sequencing (SLAF-seq) technology, and 208,993 high-quality SNPs were identified. Spray inoculation at seedling stage was used to evaluate the resistance of the 419 germplasms to 7 strains. According to phenotype and genotype data, genome-wide association study (GWAS) for rice blast was performed using general linear model (GLM) and mixed linear model (MLM). A total of 20 loci were detected under the two models, including 20 loci detected by GLM and 1 locus detected by MLM. Chr12_10803913 locus was detected in both models. There were 17 loci, overlapping with previously reported genes/QTLs, while the remaining three loci were the first reported, including Chr3_18302718, Chr3_18302744, and Chr5_10379127. A total of 323 candidate genes were screened out in the genomic regions of 150 kb upstream and downstream of 20 significantly associated loci. Eight candidate genes were preliminarily determined to be related to disease resistance. Among them, both()and() were known cloned genes,,,andwere selected as candidate genes near the three loci. The results provided the scientific basis for the mining of rice blast resistance loci and gene cloning.
rice; blast disease; genome-wide association study (GWAS); candidate genes
10.3724/SP.J.1006.2021.02047
本研究由中央引導地方科技發展專項(桂科ZY19183020), 廣西創新驅動發展專項(AA17204045-1), 廣西自然科學基金項目(2020GXNSFAA259041, 2018GXNSFAA138124, 2017GXNSFBA198210), 廣西重大科技創新基地開放課題(2018-05-Z06-CX04)和廣西農業科學院發展基金(桂農科2019Z08)資助。
This study was supported by the Special Fund of Local Science and Technology Development for the Central Guidance (Guike ZY19183020), the Guangxi Special Fund for Innovation-Driven Development (AA17204045-1), the Guangxi Natural Science Fund (2020GXNSFAA259041, 2018GXNSFAA138124, 2017GXNSFBA198210), the Opening Project of Major Science and Technology Innovation Base for Guangxi (2018-05-Z06-CX04), and the Development Fund of Guangxi Academy of Agricultural Sciences (Guinongke 2019Z08).
李丹婷, E-mail: ricegl@163.com; 楊行海, E-mail: yangxinghai514@163.com
**同等貢獻(Contributed equally to this work)
陳燦, E-mail: chencan129@126.com; 農保選, E-mail: nongbaoxuan88@gxaas.net
2020-07-12;
2020-12-01;
2020-12-28.
URL: https://kns.cnki.net/kcms/detail/11.1809.S.20201228.1435.008.html

