ceRNA對植物纖維素形成的調控研究進展
詹妮,謝耀堅,吳志華,劉果,尚秀華
?
ceRNA對植物纖維素形成的調控研究進展
詹妮,謝耀堅,吳志華*,劉果,尚秀華
(國家林業和草原局桉樹研究開發中心,廣東 湛江 524022)
植物纖維素的形成是由多個基因參與且呈網絡調控。通過對纖維素形成過程中的關鍵酶基因、轉錄因子以及ceRNA研究的闡述,深入了解纖維素生物合成調控機制。綜述植物纖維素形成過程中的纖維素合酶、蔗糖合成酶、MYB等重要基因以及lncRNA、miRNA、circRNA類ceRNA,闡述其復雜的分子調控網絡,以期解析植物纖維素形成過程中的分子調控機制,深入了解植物纖維素形成過程。
競爭性內源RNA;纖維素;轉錄因子;表達調控
纖維素作為植物細胞壁中必不可少的結構成分,發揮著重要的作用。植物細胞初生壁中的纖維素微纖絲在植物細胞的擴增階段調控植物形態建成,次生壁中的纖維素使植物細胞具有特定功能,纖維素對植物生長的重要作用使得對其研究具有重要意義[1]。纖維素形成是個較復雜的過程,該過程涉及一系列重要生物學過程,其中每個過程均由多基因參與且呈網絡調控,研究植物纖維素形成過程中的關鍵基因及其生物合成調控機制,已成為當前研究的熱點[2]。
ceRNA(Competing endogenous RNA,競爭性內源RNA)是指生物體內復雜的轉錄調控網絡中的RNA,包括長鏈非編碼RNA(Long noncoding RNA,lncRNA)、微小RNA(MicroRNA,miRNA)以及環狀RNA(CircleRNA,circRNA)等[3-4]。ceRNA通過miRNA應答元件(MicroRNA response element,MRE)與靶mRNA競爭性的結合同種的miRNA分子,使miRNA的表達水平及活性相對下降,從而抑制了miRNA對靶mRNA 的沉默效應,發揮調控的作用[5-7]。RNA轉錄物通過它們所共有的 MREs位點來彼此調節,共有的MREs數量越多,它們的交流或共調節的程度也越大,因此ceRNA 能形成一種大規模轉錄調控網絡,實現lncRNAs,miRNAs及circRNAs等通過MREs進行相互作用,通過競爭MREs 構成一個完整復雜的ceRNA分子調控網絡[6]。
1 植物纖維素形成過程中的關鍵基因
纖維素合酶(Cellulose synthase, CesA)組成纖維素合酶復合體(Cellulose synthase complex, CSC),催化β-1,4糖苷鍵的形成,合成纖維素,在植物纖維素合成途徑中發揮主要調節作用[8]。在大青楊()、苧麻()、馬尾松()等木本植物都相繼克隆出CesA基因[9-11]。在擬南芥()中CesA1、CesA3、CesA6 負責初生壁的形成[12-14],CesA4、CesA7、CesA8負責次生壁的合成[15-16]。
蔗糖合成酶(Sucrose synthase, SuSy)影響植物細胞分化以及細胞壁的形成,能夠提供細胞壁合成的底物,SuSy的活性與纖維素合成有關[17-23]。SuSy基因廣泛存在于植物中,CARDINI等[24]首次從小麥()中克隆了SuSy基因,此后在擬南芥、棉花(.)、馬鈴薯()、胡蘿卜()、玉米()、柑橘()、水稻()、棗()、甘蔗()等植物中獲得SuSy基因[25-31]。SuSy基因對棉花、煙草()以及楊樹的纖維素含量、纖維長度以及纖維強度等至關重要[32-34]。
擴展蛋白(Expansin,EXP)是植物細胞壁重要的組成部分,調節細胞伸展性,通過打斷細胞壁纖維素和半纖維素之間的非共價鍵,從而改變細胞壁承重網絡,使其產生位移,導致細胞壁伸展,加速細胞生長,調節組織生長[35-36]。1989年COSGROVE[37]首次從黃瓜()根尖細胞壁中提取分離出EXP。EXP能夠塑造初生細胞壁中纖維素-半纖維素網絡,EXP的活動能夠影響細胞壁的結構和組分,進而影響纖維和導管的形態[38]。GRAY等[39]在楊樹中克隆了α-EXP基因和β-EXP基因,發現PttEXP1基因在成熟莖段的次生生長較為活躍。XU等[40]發現在棉花纖維細胞的伸長過程中,α-EXP蛋白發揮了重要的調控作用。SARA等[41]從牽牛花()中獲得了一條PhEXP1基因,反義轉化后,發現牽牛花表皮細胞面積也相應的減少,細胞壁發生了改變,導致細胞壁機械強度下降。
AGO蛋白(Argonaute protein)主要包含PAZ和PIWI結構域,是小RNA介導的RNA沉默通路中RNA誘導的沉默復合物(RNA-induced silencing complex,RISC)的核心成分。AGO蛋白通過與miRNAs(microRNAs)、siRNAs(small interfering RNAs)、piRNAs(Piwi-interacting RNAs)等不同類型的小非編碼RNA(small non-coding RNA)結合,AGO蛋白能夠特異地停留在與小RNA互補的靶基因mRNA上,其自身的內切酶可以對目標靶基因進行切割,從而引起靶基因沉默,在調控植物生長發育中起到重要的作用[42-43]。
第3類亮氨酸拉鏈蛋白(ClassⅢ homeodomain leucine zipper, HD-ZipⅢ)轉錄因子、MYB(V-myb avian myeloblastosis viral oncogene homo)轉錄因子以及NAC(No apical meristem/Arabidopsis thaliana transcription activator factor/CUP- -shaped cotyledon)轉錄因子等在植物細胞生長發育過程中具重要調控作用,參與植物的生長代謝調控[44]。
研究表明,在擬南芥和水稻HD-ZipⅢI家族各有5個成員[45],毛果楊()基因組中則含有8個HD-ZipⅢ基因家族成員[46]。在白云杉()和火炬松()中已分別被克隆到4和5個HD-Zip III基因[47]。MYB類轉錄因子參與植物苯丙烷類代謝途徑的調節,調控次生細胞壁的形成[48-49]。目前,MYB轉錄因子已在擬南芥、金魚草()、大豆()、煙草、蘋果()、白樺(、毛白楊()等物種中分離并鑒定[50]。HAI 等[51]對玉米和擬南芥MYB轉錄因子分析發現,有4個亞組的MYB轉錄因子參與調控次生壁的增厚。劉慧子等[52]研究表明,白樺MYB家族中17條MYB家族基因中的絕大部分參與調控形成層的發育。葉勝龍[53]研究發現,毛白楊MYB055轉錄因子參與調控次生壁合成,影響苯丙氨酸代謝途徑,從而調控纖維素合成等相關基因的表達。
2 植物纖維素形成過程中的ceRNA
測序技術日益發展使基因數據庫與轉錄組數據庫日益充實,為ceRNA的挖掘和功能研究提供了有利的數據支持。ceRNA在生物發育和基因表達中發揮著復雜的精確調控功能,對其深入研究有助于揭示基因表達調控網絡對于生命體的復雜性[54]。
lncRNA指長度大于200個核苷酸,但含有1個少于100個氨基酸開放閱讀框(Open reading frame,ORF)的RNA,可分為長鏈非編碼自然反義轉錄本(Long noncoding natural antisense transcripts,lincNATs)、內含子 lncRNAs(Intronic lncRNAs)、啟動子 lncRNAs(Promoter lncRNAs)和長鏈基因間 ncRNAs(Long intergenic ncRNAs,lincRNAs)。lncRNAs 可作為與其互作分子的招募者、系結者、引導者、誘捕者和信號分子,從而發揮調控作用[55-57]。lncRNAs通過與miRNA結合,從而隔離miRNA,調控miRNA的表達水平,降低miRNA對mRNA的調控,最終促進了mRNA的表達。在植物中鑒定出大量lncRNAs,如擬南芥[58]、小麥[59]、玉米[60]、谷子()[61]、棉花[62]、江南卷柏()[63]、沙棘()[64]、芒草()[65]以及毛果楊、毛白楊[66-67]。
miRNA是一類內生的且長度約為20 ~ 24個核苷酸的小RNA,在轉錄以及轉錄后的過程中調控基因表達[68]。miRNA在細胞內具有多種重要的調節作用,參與了植物器官發育、代謝調節,與細胞的增殖、分化、凋亡等一系列生理過程密切相關[69]。miRNA是通過切割目標靶基因mRNA或抑制其翻譯來實現對目標靶基因的調控,這種調控既能夠通過一個miRNA調控多個基因的表達,亦可通過幾個miRNA共同調控某個基因的表達,從而形成復雜的調控網絡[70]。MCNAIR[71]研究發現12個miRNA在正常生長和快速生長桉樹()中的表達模式,miRNA在正常桉樹和應拉木的發育過程中起重要作用。利用高通量測序技術挖掘了包括2個藍桉()基因型的木質部,獲得了大量的miRNA信息[72]。李崇奇等[73]研究表明,有41個miRNA與巨桉()木質形成相關,主要調控ARF、HD-ZIPIII、KAN、MYB 和NAC轉錄因子。circRNA是一類線性閉合環狀內源性的非編碼RNA分子,circRNA通過吸附miRNA并參與其表達調控過程,circRNA 能夠特異性結合miRNA,使其失去調控mRNA 的功能,調控基因表達等生物過程[74-77]。circRNA分為外顯子circRNA、基因間circRNA和內含子circRNA[78-79]。circRNA廣泛表達于不同的植物中,表達具有時空組織特異性,circRNA作為內源性非編碼RNA在真核生物的生長發育過程中發揮著重要作用,引起人們廣泛的關注[80]。2014年在擬南芥根部發現circRNA后[81],2015年ANDREEVA和COOPER研究報道了circRNA廣泛存在于動植物細胞組織中,且具有很多特殊的生物學特性之后,引起國內外科學家的高度重視[82]。在水稻[83-84]、大麥()[85]、番茄()[86]以及小麥[87]中發現存在大量的circRNAs。YE等[84]在水稻的根和擬南芥的葉中分別鑒定了12 037和6 012個circRNAs。
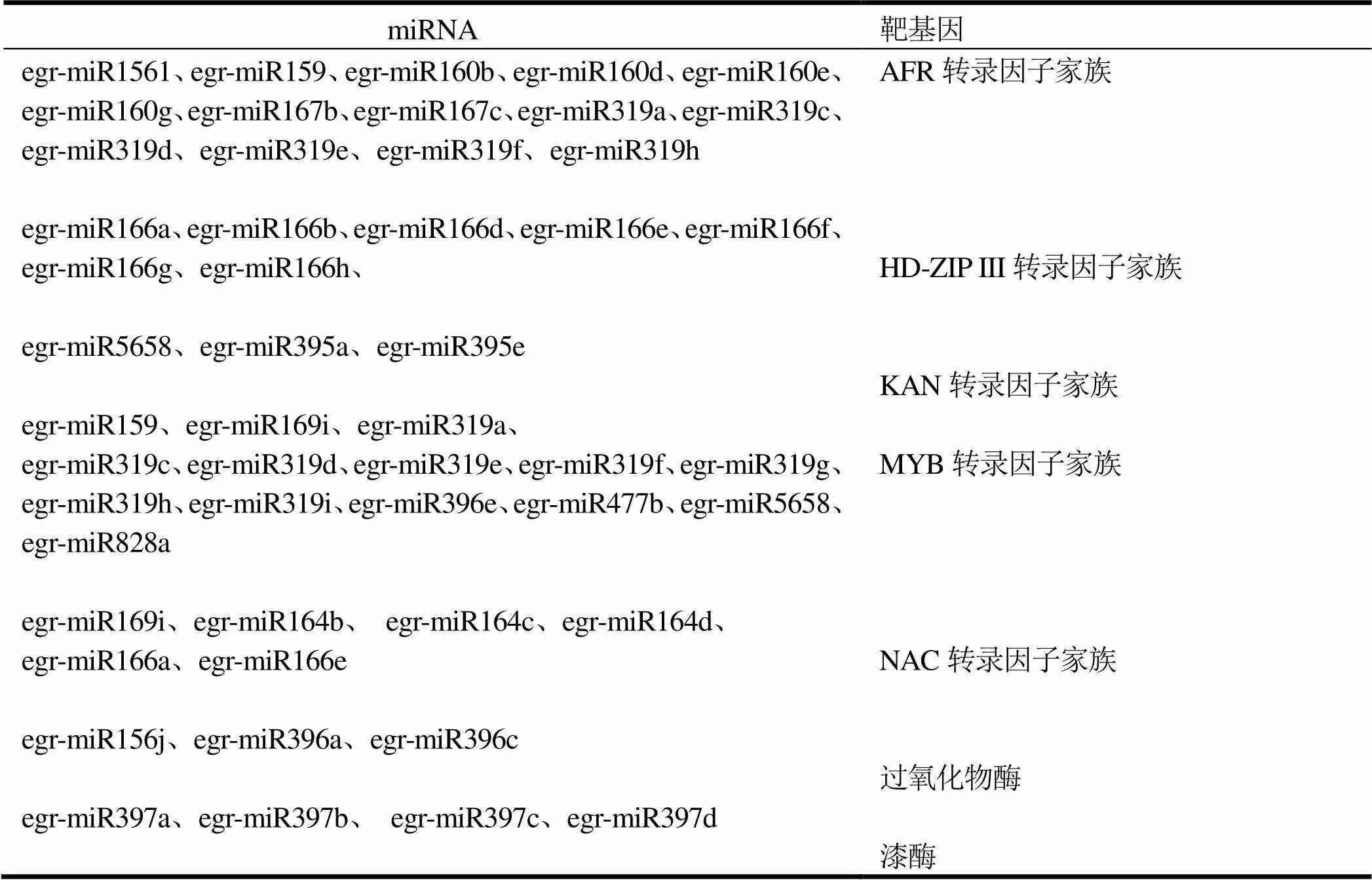
表1 與木質相關的miRNA[73]
通過對參與植物纖維素形成過程中的關鍵基因、ceRNA的進一步研究,能夠更好的解析植物纖維素形成過程中的分子調控機制,以期獲得對植物纖維素形成過程的深入了解,從而為育種工作服務。現今在植物中已經鑒定出許多ceRNA,但只有少數做了功能驗證,今后植物ceRNA的研究方向可能會趨向于搜索基因的功能,剖析功能冗余以及其應用等方面。
[1] SETHAPPHONG L, DAVIS J K, SLABAUGH E, et al. Prediction of the structures of the plant specific regions of vascular plant cellulose synthases and correlated functional analysis[J]. Cellulose, 2016, 23 (1): 145-161.
[2] PLOMION C, LEPROVOST G, STOKES A. Wood formation in trees[J]. Plant Physiology, 2001, 127 (4): 1513-1523.
[3] STECK E. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions[J]. Journal of Molecular Medicine, 2012, 90(10):1185-1195.
[4] CHANG H Y. Long noncoding RNAs: cellular address codes in development and disease[J]. Cell, 2013, 152(6):1298-1307.
[5] RINN J L, CHANG H Y. Genome regulation by long noncoding RNAs[J]. Annual Review of Biochemistry, 2012, 81(1): 145-166.
[6] SALMENA L, POLISENO L, TAY Y, et al. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language?[J]. Cell, 2011, 146(3):353-358.
[7] BARTEL D P. MicroRNAs:Target recognition and regulatory functions[J]. Cell, 2009, 136(2): 215-233.
[8] GU Y, KAPLINSKY N, BRINGMANN M, et al. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(29): 12866-12871.
[9] 許雷,劉一星,方連玉.大青楊纖維素合成酶 PuCesA6 基因cDNA 的克隆及序列分析[J].西南林業大學學報,2012,32(5):26-32.
[10] 劉昱翔,陳建榮,彭彥,等.苧麻纖維素合成酶基因BnCesA4 cDNA 序列的克隆與表達分析[J].作物研究, 2014,28(5):472-478.
[11] 阮維程,潘婷,季孔庶.馬尾松纖維素合成酶基因PmCesA1的克隆及其分析[J].分子植物育種,2015,13(4):861-870.
[12] ARIOLI T, PENG L C, BETZNER A S, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis[J]. Science, 1998, 279 (5351): 717-720.
[13] FAGARD M, DESNOS T, DESPREZ T, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis[J]. Plant Cell, 2000, 12(12): 2409-2424.
[14] SCHEIBLE W R, ESHED R, RICHMOND T, et al. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants[J]. Proceedings of the National Academy of Sciences of the United States of America,2001,98(18): 10079-10084.
[15] TAYLOR N G, LAURIE S, TURNER S R. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis[J]. Plant Cell, 2000, 12(12): 2529-2539.
[16] TURNER S R, SOMERVILLE C R. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall[J].Plant Cell, 1997, 9(5): 689-701.
[17] BARRERO S C, HERNANDO A S, GONZALEZ M P, et al. Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley[J]. Planta, 2011, 234(2): 391-403.
[18] POOVAIAH C R, MAZAREI M, DECKER S R, et al. Transgenic switchgrass(L.) biomass is increased by over-expression of switchgrass sucrose synthase (PvSUS1)[J]. Biotechnology Journal, 2015, 10(4): 552-563.
[19] 房經貴,朱旭東,賈海鋒,等.植物蔗糖合酶生理功能研究進展[J].南京農業大學學報,2017,40(5):759-768.
[20] COLEMAN H D, YAN J, MANSFIELD S D, et al. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(31):13118-13123.
[21] FUJII S, HAYASHI T, MIZUNO K. Sucrose synthase is an integral component of the cellulose synthesis machinery[J]. Plant and Cell Physiology, 2010, 51(2): 294–301.
[22] BAI W Q, XIAO Y H, ZHAO J, et al. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers[J]. Plos One, 2014, 9(5): e96537.
[23] 詹妮,謝耀堅,陳鴻鵬,等.巨桉 SuSy 基因家族的生物信息學分析[J].熱帶亞熱帶植物學報,2018,26(6):580-588.
[24] CARDINI C E, LELOIR F, CHIRIBOGA J. The biosynthesis of sucrose [J]. Journal of Biological Chemistry, 1955, 214(1): 149–156.
[25] 雷美華,葉冰瑩,王冰梅,等.甘蔗蔗糖合成酶基因的克隆[J].應用與環境生物學報,2008,14(2):177-179.
[26] BAUD S, VAULTIER M N, ROCHAT C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis[J]. Journal of Experimental Botany, 2004, 55(396): 397-409.
[27] CARLSON S J, CHOUREY P S, HELENTJARID T, et al. Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene[J]. Plant Molecular Biology, 2002, 49(1): 15–29.
[28] HIROSE T, SCOFIELD G N, TERAO T. An expression analysis profile for the entire sucrose synthase gene family in rice[J]. Plant Science, 2008, 174(5): 534-543.
[29] CHEN A Q, HE S, LI F F, et al. Analyses of the sucrose synthase gene family in cotton: Structure, phylogeny and expression patterns[J]. BMC Plant Biology, 2012, 12(1): 85-102.
[30] 馮延芝,魏琦琦,何瀟等.棗蔗糖合成酶基因SS6的克隆及表達分析[J].經濟林研究,2017(4):36-42.
[31] 賈春平,耿洪偉,朱亞夫,等.海島棉蔗糖合成酶基因克隆及生物信息學和表達模式分析[J].分子植物育種,2016,14(2):286-301.
[32] JIANG Y J, GUO W Z, ZHU H Y, et al. Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality[J]. Plant Biotechnology Journal, 2012, 10(3): 301-312.
[33] WEI Z G, QU Z S, ZHANG L J, et al. Overexpression of poplar xylem sucrose synthase in tobacco leads to a thickened cell wall and increased height[J]. Plos One, 2015, 10(3): e0120669.
[34] XU S M, BRILL E, LLEWELLYN D J., et al. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production[J]. Molecular Plant, 2012, 5(2): 430-441.
[35] 趙美榮.植物擴展蛋白基因及其表達調控的研究進展[J].赤峰學院學報(自然版),2014(14):1-5.
[36] COSGROVE D J. Growth of the plant cell wall[J]. Nature Reviews Molecular Cell Biology, 2005, 6(11): 850-861.
[37] COSGROVE D J. Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls[J]. Planta, 1989, 177(1): 121-130.
[38] GRAY M M, BLOMQUIST K, MCQUEEN M S, et al. Ectopic expression of a wood-abundant expansin PttEXPA1 promotes cell expansion in primary and secondary tissues in aspen[J]. Plant Biotechnology Journal, 2008, 6(1): 62-72.
[39] GRAY M M, MELLEROWICZ E J, ABE H, et al. Expansins abundant in secondary xylem belong to subgroup A of the alpha-expansin gene family[J]. Plant Physiology, 2004, 135(3):1552-1564.
[40] XU B, GOU J Y, LI F G, et al. A cotton BURP domain protein interacts with α-Expansin and their Co-expression promotes plant growth and fruit production[J]. Molecular Plant, 2013, 6(3): 945-958.
[41] SARA Z, LARA R, GIOVANNI B T, et al. Down regulation of the Petunia hybrida α-Expansin Gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in Petal Limbs[J]. Plant Cell, 2004, 16(2): 295-308.
[42] CENIK E S, ZAMORE P D. Argonaute proteins[J]. Current Biology, 2011,21(12):446-449.
[43] 范春節,閆慧芳,裘珍飛,等.巨桉AGO基因家族的生物信息學分析[J].熱帶亞熱帶植物學報,2015,23(4): 361-369.
[44] 程健弘,魏明科,林二培,等.杉木HD-ZipⅢ轉錄因子的克隆及表達分析[J].農業生物技術學報,2017(11):1820-1830.
[45] ITOH J I, HIBARA K I, SATO Y, et al. Developmental Role and Auxin Responsiveness of Class III Homeodomain Leucine Zipper Gene Family Members in Rice[J]. Plant Physiology, 2008, 147(4):1960-1975.
[46] KO J H, PRASSINOS C, HAN K H. Developmental and seasonal expression of PtaHB1, agene encoding a class III HD-Zip protein, is closely associated with secondary growth and inversely correlated with the level of microRNA (miR166)[J]. New Phytologist, 2006, 169(3): 469-478.
[47] CAROLINE L, COTEET B F, ROY V, et al. Gene family structure, expression and functional analysis of HD-Zip III genes in angiosperm and gymnosperm forest trees[J]. BMC Plant Biology, 2010, 10(1): 273.
[48] GEETHALAKSHMI S, BARATHKUMAR S, PRABU G. Thetranscription factor family genes in Sugarcane () [J].Plant Molecular Biology Reporter, 2015, 33(3):512-531.
[49] LI C, WANG X, RAN L, et al.PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation intomentosa[J]. Plant and Cell Physiology, 2015, 56(12): 24-36.
[50] LI C, NG C K Y, FAN L M. MYB transcription factors, active players in abiotic stress signaling[J]. Environmental & Experimental Botany, 2015, 114:80-91.
[51] HAI D, BO R F, SI S Y, et al. The R2R3-MYB transcription factor gene family in Maize[J]. Plos One, 2012, 7(6):e37463.
[52] 劉慧子,孫丹,于穎,等.白樺MYB家族基因序列及表達分析[J].植物研究,2016,36(2):252-257.
[53] 葉勝龍.毛白楊MYB055轉錄因子在次生壁合成中的調控機制研究[D].重慶:西南大學,2015.
[54] SARVER A L, SUBRAMANIAN S. Competing endogenous RNA database[J]. Bioinformation, 2012, 8(15):731-733.
[55] ZHANG Y, TAO Y, LIAO Q. Long noncoding RNA: a crosslink in biological regulatory network[J]. Briefings in Bioinformatics, 2018,19(5):930-945..
[56] ANITA Q G, SOFIA N, HELENA S. Non-Coding RNAs: multi-tasking molecules in the cell[J].International Journal of Molecular Sciences, 2013, 14(8):16010-16039.
[57] WILUSZ J E. Long noncoding RNAs: re-writing dogmas of RNA processing and stability[J]. Biochimica et Biophysica Acta-biomembranes,2016,1859(1): 128-138.
[58] WANG H, CHUNG P J, LIU J, et al. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis[J]. Genome Research, 2014, 24(3):444-453.
[59] CAGIRICI H B,ALPTEKIN B,BUDAK H.RNA sequencing and co-expressed long non-coding RNA in modern and wild wheats[J].Scientific Reports,2017,7(1): 10670.
[60] LI L, EICHTEN S R, SHIMIZU R, et al. Genome-wide discovery and characterization of maize long non-coding RNAs[J]. Genome Biology, 2014, 15(2): R40.
[61] QI X, XIE S, LIU Y, et al. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing[J].Plant Molecular Biology, 2013, 83(4-5): 459-473.
[62] LU X, CHEN X, MU M, et al. Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton(L.)[J]. Plos One, 2016, 11(6): e0156723.
[63] ZHU Y, CHEN L, ZHANG C, et al. Global transcriptome
analysis reveals extensive gene remodeling, alternative splicing and differential transcription profiles in non-seed vascular plant Selaginella moellendorffii[J].BMC Genomics, 2017, 18(Suppl1): 1042.
[64] ZHANG G, DUAN A, ZHANG J, et al. Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn(Linn)fruit[J]. Gene, 2017, 596: 130-136.
[65] XU Q, SONG Z, ZHU C, et al. Systematic comparison of lncRNAs with protein coding mRNAs in population expression and their response to environmental change[J]. BMC Plant Biology, 2017,17(1): 42.
[66] LV Y, LIANG Z, MIN G, et al. Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize(L.)[J].BMC Genomics, 2016, 17(1): 350.
[67] TIAN J, SONG Y, DU Q, et al. Population genomic analysis of gibberellin-responsive long non-coding RNAs in[J]. Journal of Experimental Botany, 2016, 67(8):2467-2482.
[68] AXTELL M J. Classification and comparison of small RNAs from plants[J].Annual Review Plant Biology,2013, 64:137-159.
[69] PARK S M, PETER M E. microRNAs and death receptors[J]. Cytokine & Growth Factor Reviews, 2008, 19(3/4): 303-311.
[70] SHUAI P, LIANG D, ZHANG Z,et al.Identification of droughtresponsive and novelmicroRNAs by highthroughput sequencing and their targets using degradome analysis[J].BMC Genomics,2013,14:233.
[71] MCNAIR G R. Whole-tree and tension wood-associated expression profiles of micrornas intrees[D]. Pretoria: University of Pretoria,2009.
[72] PAPPAS M, REIS A, FARINELL L, et al. Interspecific discovery and expression profiling ofmicro RNAs by deep sequencing[J]. BMC Proceedings, 2011, 5(7):1-2.
[73] 李崇奇,沈文濤,言普,等.巨桉miRNA及其靶基因生物信息學預測[J].南方農業學報,2014,45(9):1532-1538.
[74] NIGRO J M, CHO K R, FEARON E R., et al. Scrambled exons.[J]. Cell, 2016, 64(3):607-613.
[75] EBERT M S, NEILSON J R, SHARP P A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells[J]. Nature Methods, 2007, 4(9): 721-726.
[76] 岳慧芳,任永哲,王志強,等.circRNAs在植物中的研究進展[J].西北植物學報,2018,38(2):386-392.
[77] LIU Q, ZHANG X, HU X, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation[J]. Scientific Reports, 2016, 6: 22572.
[78] CHEN B, HUANG S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer[J]. Cancer Letters, 2018, 418(18):30033-30038.
[79] GREENE J, BAIRD A M, BRADY L, et al. Circular RNAs: biogenesis, function and role in human diseases[J]. Frontiers in Molecular Biosciences, 2017, 4:38.
[80] LEE S M, KONG H G, RYU C M. Are Circular RNAs New Kids on the Block?[J]. Trends in Plant Science, 2017, 22(5):357-360.
[81] WANG P L, BAO Y, YEE M C, et al. Circular RNA Is expressed across the eukaryotic tree of life[J]. Plos One, 2014, 9(6):e90859.
[82] ANDREEVA K, COOPER N G F. Circular RNAs: new players in gene regulation[J]. Advances in Bioscience & Biotechnology, 2015, 6(6): 433-441.
[83] CHU Y Y, LI C, CHEN L, et al. Widespread noncoding circular RNAs in plants[J]. New Phytologist, 2015, 208(1): 88-95.
[84] YE C Y, ZHANG X, CHU Q, et al. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice[J]. RNA Biology, 2016, 14(8):1-9.
[85] DARBANI B, NOEPARVAR S, BORG S. Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley[J]. Frontiers in Plant Science, 2016, 7:776.
[86] ZUO J, WANG Q, ZHU B, et al. Deciphering the roles of circRNAs on chilling injury in tomato[J]. Biochemical & Biophysical Research Communications, 2016, 479(2): 132-138.
[87] WANG Y, YANG M, WEI S, et al.Identification of circular RNAs and their targets in leaves ofL. under dehydration stress[J]. Frontiers in Plant Science, 2017, 7: 1-10.
Research Progress in the Regulation of ceRNA on Plant Cellulose Formation
ZHAN Ni, XIE Yaojian, WU Zhihua, LIU Guo, SHANG Xiuhua
(,)
The formation of plant cellulose is regulated by multiple genes and pathways. In this paper, the key enzyme genes, transcription factors and ceRNA in the process of cellulose formation are elaborated to further understand the regulation mechanism of cellulose biosynthesis. The key genes including cellulose synthase, sucrose synthase, MYB and ceRNA including lncRNA, miRNA and circRNA in the process of plant cellulose formation are reviewed. The complex molecular control network is expounded in order to analyze the molecular control mechanism of plant cellulose formation, and to understand the process of plant cellulose formation.
ceRNA; cellulose; transcription factors; expression regulation
Q74
A
國家自然科學基金面上項目“桉樹抗風特性及其主要影響因子研究”(31570615);國家重點研發計劃課題“桉樹、云南松(思茅松)、華山松豐產增效技術集成與示范”(2017YFD0601202)
詹妮(1990― ),女,博士研究生,主要從事桉樹林木遺傳育種方面的研究,E-mail: jennyzn1122@163.com
吳志華(1974― ),男,副研究員,主要從事林木逆境生理研究,E-mail: wzhua2889@163.com

