植物低溫脅迫應答bHLH轉錄因子研究進展



摘要:低溫環境嚴重限制了植物的生長發育和作物產量。在長期的進化過程中,植物在分子、生理和生化水平上形成了復雜的適應低溫脅迫的機制。其中,在分子水平上,轉錄因子是低溫脅迫信號傳導的主要參與者,一些轉錄因子構成了信號網絡中的主要樞紐。該網絡中主要的轉錄因子包括MYB、bHLH、bZIP、ERF、NAC和WRKY等。人們利用基因工程、生物信息學和轉錄組學等多種生物學技術,逐漸發現了bHLH轉錄因子在植物逆境響應中的重要作用。bHLH轉錄因子在低溫脅迫響應中起關鍵作用,是植物分子育種中提高低溫脅迫耐受性的寶貴基因資源。因此,闡明bHLH轉錄因子植物響應低溫脅迫的調控機制具有重要意義。本文簡要介紹了bHLH轉錄因子的基本特征和各植物中bHLH家族成員數量,對參與低溫脅迫的bHLH轉錄因子家族成員(如ICE、MYC2)進行敘述,并闡述了bHLH轉錄因子在低溫脅迫中的調控機制,包括與下游靶基因或其他轉錄因子互作、ICE-CBF通路和活性氧(ROS)及植物激素介導的信號通路,為進一步提高植物抗寒性和培育抗寒新品種提供參考。
關鍵詞:bHLH轉錄因子;低溫脅迫;調控;逆境響應;抗寒性
中圖分類號:S184 文獻標志碼:A
文章編號:1002-1302(2024)16-0011-09
低溫限制了冷敏感植物的生長發育、地理分布、產量和品質,對植物的影響顯著大于其他環境脅迫因素[1]。低溫脅迫對植物的影響主要表現在光合作用減弱、水分代謝失調、酶活性降低,對植物生長和產量造成不可逆的傷害[2]。為了在惡劣條件下生存,許多植物通過轉錄因子調控一系列靶基因的表達來增強植物對低溫脅迫的適應性[3]。轉錄因子根據DNA結合區域的特異性可分為多個不同的家族,與植物抗逆性相關的主要有4種:堿性亮氨酸拉鏈(bZIP)、WRKY、AP2/ERF(APETALA2/ethylene responsive factor)、MYB(V-myb avian myeloblastosis viral oncogene homolog)。堿性螺旋-環-螺旋(bHLH)轉錄因子家族在抗逆研究中相對滯后[4]。近年來隨著對bHLH轉錄因子家族研究的不斷深入,發現其在抵御低溫脅迫中有重要的調控作用,比如擬南芥(Arabidopsis thaliana)中的ICE1(inducer of CBF expression 1)和ICE2(inducer of CBF expression 2)、野生稻(Oryza rufipogon)中的OrbHLH001參與低溫脅迫應答,并可構成調控網絡[5-7]。本文重點綜述了bHLH轉錄因子在調控植物低溫脅迫耐受性分子機制中的作用,這些研究為提高植物的抗寒性奠定了基礎。
1 bHLH轉錄因子的結構特征與分類
bHLH家族是真核生物轉錄因子中僅次于MYB的第二大家族,廣泛存在于植物中,其命名來源于其具有高度保守的堿性/螺旋-環-螺旋特殊結構域[8]。它由2個部分組成,一部分是堿性氨基酸區域,另一部分是螺旋-環-螺旋區域(HLH)(圖1)。堿性結構域位于bHLH的N端,包含約15個氨基酸和6個堿性殘基,其主要參與DNA與靶基因中的E-box(CANNTG)或G-box(CACGTG)基序的結合[9];HLH區域位于C端,包含40~50個氨基酸,包括2個由疏水殘基通過疏水環連接的兩親性α-螺旋。由于α-螺旋具有靈活性,可以進行結構域二聚化以促進蛋白質-蛋白質相互作用,并形成同源或異源二聚體復合物來調控靶基因的表達[10]。
bHLH家族首先在小鼠肌肉發育研究中被發現,隨后在所有真核生物包括動物、植物和真菌中被鑒定[11-12]。1989年,Ludwig等在玉米中發現了由R(resistance)基因編碼花青素合成的第1種植物
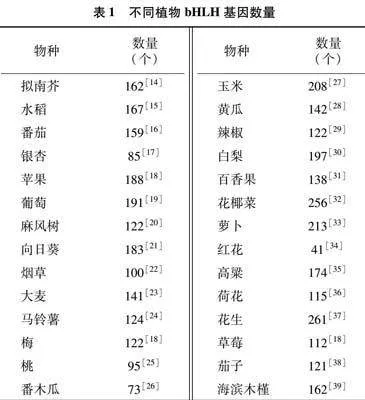
bHLH蛋白[13]。bHLH家族基因的數量在不同的物種之間也有差異(表1):在擬南芥(Arabidopsis thaliana)的基因組中共鑒定出162個基因(AtbHLH)[14],而在水稻(Oryza sativa)中發現了167個(OsbHLH),在番茄(Solanum lycopersicum)中發現了159個(SlbHLH),在銀杏(Ginkgo biloba)中發現了85個(GbbHLH),在蘋果(Malus domestica)中發現了188個(MdbHLH),在葡萄(Vitis vinifera)中發現了191個(VvbHLH)[15-19]。
1999年,Atchley等根據進化關系、與DNA的結合模式以及自身的功能特性,首次將動物bHLH分為6個亞家族(A~F)[9]。A組可以特異性結合 E-box 核心序列;B組成員可與具有CACGTG或CATGTTG特征的E-box結合;C組可以結合ACGTG或GCGTG序列;D組成員沒有基本的DNA結合區域,主要參與與其他bHLH家族蛋白的異源二聚化;E組可以優先與具有CACGCG或CACGAG堿基的 N-box 結合;而F組成員可以結合特定的DNA靶序列,僅有1個COE家族[40]。擬南芥中首次構建了植物bHLH的系統進化樹,擬南芥bHLH家族被分為12個亞家族[41]。在植物的發育過程中,某些亞家族可能起著重要的調節生物反應的作用。
2 與低溫脅迫有關的bHLH轉錄因子
近年來,一些研究表明,bHLH轉錄因子參與冷脅迫響應過程并發揮重要作用。ICE1編碼一個典型的MYC-like堿性螺旋-環-螺旋(bHLH)轉錄因子,是第1個被鑒定的CBF[C-repeat-binding factors,又稱DREB(dehybration responsive element binding protein)]上游調控因子,在冷信號傳導途徑中起關鍵作用[5]。Yang等從南亞熱帶果樹龍眼(Dimocarpus longan)中分離得到1個新的ICE-like基因DlICE1,過表達DlICE1的轉基因擬南芥的AtCBF1/AtCBF2/AtCBF3基因和冷響應基因(AtRD29a、AtCOR15A、AtCOR47和AtKIN1)的表達水平比野生型的更高,并通過增加脯氨酸(Pro)含量、降低離子滲漏、減少丙二醛(MDA)和活性氧(ROS)積累來增強耐冷性[42]。Wang等研究發現,StICE1在擬南芥中的過表達增強了活性氧的清除能力及CBFs和COR(cold-related)基因的表達水平。并且StICE1通過結合StLTI6A基因的啟動子,從而維持細胞膜的穩定性,并增強了對冷脅迫的耐受性[43]。在低溫脅迫下,植物過表達SlICE1a增強了CBF/DREB及其靶基因的誘導,增加了脯氨酸、可溶性糖的水平,從而增強了煙草(Nicotiana tabacum)對冷脅迫的耐受性[44]。Zuo等的研究表明,過表達日本結縷草(Zoysia japonica)ZjICE1的轉基因擬南芥表現出對冷脅迫的耐受性增強,SOD、POD活性增加,游離脯氨酸含量增加,MDA含量降低[45]。Man等研究發現,異源表達蘿卜(Raphanus sativus)RsICE1基因提高了水稻對低溫脅迫的耐受性,表現為較高的存活率、較高的可溶性糖和游離脯氨酸含量、較低的電解質滲透率和MDA含量以及較高的葉綠素含量[46]。ICE2是ICE1的旁系同源基因,也具有類似的功能。Zhang等從切花菊神馬(Chrysanthemum morifolium ‘Jinba’)中鑒定出ICE家族基因CmICE2,過表達CmICE2的擬南芥植株比野生型更耐凍,并且經過-9 ℃低溫處理6 h后,CmICE2過表達植株中AtCBF1、AtCBF2、AtCBF4、AtCOR6.6A、AtCOR414和AtKIN1等基因的表達量也顯著上調;過表達CmICE2的植株脯氨酸含量及超氧化物歧化酶(SOD)、過氧化物酶(POD)和過氧化氫酶(CAT)的活性也顯著提高[47]。異源表達來自野生山葡萄(Vitis amurensis)的2個bHLH基因VaICE1和VaICE2的轉基因擬南芥植株耐冷性增強[48]。Zuo等從日本結縷草中鑒定得到1個新的MYC型bHLH轉錄因子ZjICE2,在擬南芥中過表達ZjICE2增強了其對冷、干旱和鹽脅迫的耐受性[49]。Fursova等從擬南芥中鑒定得到1個新的正調控低溫脅迫的bHLH家族轉錄因子ICE2,在轉基因擬南芥植株中過表達ICE2,冷馴化后對低溫脅迫的耐受性增強。過表達ICE2的轉基因株系的種子表現為碳水化合物水平降低和脂質水平升高[6]。
除ICE蛋白外,bHLH轉錄因子家族的其他一些成員也參與植物的低溫脅迫。煙草NtbHLH123是一種轉錄激活因子,可與NtCBF基因啟動子中的G-box/E-box基序結合,調控ROS清除相關和脅迫響應基因的表達,從而提高抗寒性[50]。Wang等研究發現,在王林蘋果(Malus domestica ‘Orin’)愈傷組織中過表達MdMYC2提高了MdCIbHLH1、MdCBF1、MdCBF2和MdCBF3的表達水平,從而提高了其抗凍性[51]。Wang等研究發現,TaMYC2在低溫脅迫響應中起正調控作用。過表達TaMYC2提高了擬南芥抗凍性,包括積累脯氨酸含量和增加ROS清除活性[52]。從我國野生山葡萄耐寒種質黑龍江實生苗中鑒定得到的葡萄bHLH轉錄因子VabHLH1和歐洲葡萄赤霞珠(V. vinifera ‘Cabernet Sauvignon’)中鑒定得到的bHLH轉錄因子VvbHLH1作為轉錄激活因子參與了低溫脅迫。在轉基因擬南芥植株中過表達VabHLH1和VvbHLH1轉錄因子不影響轉基因擬南芥的生長發育,但增強了其對低溫脅迫的耐受性[53]。異源表達MdCIbHLH1基因增強了轉基因擬南芥和轉基因煙草的耐冷性[54]。在轉基因煙草中過表達秋子梨(Pyrus ussuriensis)PubHLH1,增強了對冷脅迫的耐受性。過表達PubHLH1的轉基因株系表現為較高的存活率、較高的葉綠素含量和脯氨酸含量、較低的電解質滲透率和MDA含量[55]。苦蕎麥(Fagopyrum tataricum)FtbHLH2是一個冷相關轉錄因子,在增強冷脅迫耐受性中發揮正調控作用。在擬南芥中過表達FtbHLH2通過上調相關酶的表達,提高轉基因植株對低溫脅迫的抗性[56]。辣椒(Capsicum annuum)CabHLH79通過調控抗氧化系統酶(SOD、POD和CAT)和抗寒相關基因(AtRD29a、AtERD15和AtCBF1)的表達來增強抗寒性[57]。
最新研究發現,屬于bHLH轉錄因子家族Ⅶ類轉錄因子的光敏色素互作因子PIFs也參與了植物的低溫脅迫。Lee等研究發現,CBF途徑會受到光周期的調控,黎明后8 h,短日照(SD)植株的CBF轉錄水平是長日照(LD)植株的3~5倍,SD植株的抗凍性大于LD植株,并且光敏色素B(PHYB)和2個光敏色素互作因子PIF4和PIF7在低溫脅迫下下調了CBF途徑和耐凍性[58]。Jiang等研究發現,PIF3通過直接結合CBF基因的啟動子來下調其表達,從而作為擬南芥抗凍性的負調節因子發揮作用[59]。甜橙(Citrus sinensis)中一個光敏色素互作轉錄因子(PIF)基因(CsPIF8)在低溫脅迫下顯著上調,過表達CsPIF8提高了轉基因番茄植株和轉基因葡萄柚愈傷組織的耐冷性[60]。SlPIF4通過直接結合SlCBF基因的啟動子并激活其表達來正向調控番茄的耐冷性[61]。Cordeiro等在水稻中鑒定了一種光敏色素互作因子OsPIF14,它與OsDREB1B啟動子結合,下調了水稻原生質體中OsDREB1B的表達[62]。He等通過ChIP-qPCR分析、酵母雙雜交試驗、表達模式分析等試驗發現,光敏色素(phyB)缺失通過OsPIL16正向調節OsDREB1表達以增強細胞膜完整性并降低丙二醛濃度,從而提高phyB突變體的耐寒性[63]。還有其他的bHLH轉錄因子參與調控低溫脅迫,詳見表2。
3 bHLH轉錄因子在植物低溫脅迫中的調控機制
3.1 與下游靶基因或其他轉錄因子互作
bHLH轉錄因子是一類含有保守的bHLH結構域的蛋白質家族,是參與DNA結合的基序。bHLH蛋白在這些基因的啟動子區域通過序列特異性相互作用調控下游基因[91]。在擬南芥中AtICE1/AtbHLH116[JP+1]蛋白在低溫環境下與CBF啟動子區域結合影響轉錄起始,過表達AtICE1/AtbHLH116的植株對低溫表現出更高的耐受性[5]。而Hu等研究發現,AtJAZ1和AtJAZ4可以與AtICE1/AtICE2互作,抑制AtICE的穩定性及其下游CBF的表達,從而負調控擬南芥耐冷性[92]。Wang等研究發現,TaMYC2與TaICE41互作激活下游CBF-COR途徑,從而提高植物的抗凍性。TaJAZ7可能與TaMYC2互作抑制TaICE41的轉錄活性或直接干擾TaICE41抑制TaICE41基因的表達,從而負調控植物耐凍性[52]。PuICE1可以通過與PuHP1互作提高PuDREBa的轉錄表達,從而提高秋子梨的抗寒性[72]。PtrMYC2與PtrBADH-I互作可以提高甘氨酸和甜菜堿的含量,從而提高枳(Poncirus trifoliata)的耐冷性[81]。Feng等通過EMSA和ChIP-PCR驗證了MdCIbHLH1蛋白與AtCBF3和MdCBF2基因啟動子中的MYC識別序列特異性結合[54]。Xie等在探究MdbHLH3調控低溫誘導的蘋果花青素積累和果實著色的分子機制中,進行了酵母雙雜交試驗和雙分子熒光互補試驗,發現MdbHLH3與MdMYB1之間存在特異性互作關系[66]。
3.2 ICE-CBF通路
為了快速感知和響應低溫脅迫,植物進化出一系列復雜且精確的生理、生化和分子機制來響應這種脅迫。其中,依賴于CBF的途徑是植物中最重要的冷信號途徑。CBF又稱脫水應答元件結合因子1(DREB1),屬于AP2/EREBP轉錄因子家族,是冷信號途徑中的核心轉錄因子[93]。CBF基因的表達受冷脅迫快速誘導,與COR基因啟動子中的脫水應答元件(CRT/DRE)結合,激活COR基因表達,增強植物的抗寒性[5]。CBF受多個轉錄因子調控,其中CBF誘導因子(inducer of CBF,ICE)被認為是CBF的主要調控因子。ICE1與CBF啟動子中的MYC結合位點(CANNTG)結合。ICE1蛋白的功能主要受翻譯后修飾(post-translational modification,PTM)的調控[42]。ICE1蛋白中的PTM主要包括泛素化、SUMO化和磷酸化。編碼一個RING型泛素E3連接酶的HOS1(high expression of osmotically responsive genes 1)直接與ICE1蛋白相互作用并多聚泛素化導致該蛋白通過26S蛋白酶體途徑(26S proteasome pathway)降解[94]。SUMO E3連接酶SIZ1介導ICE1的SUMO化修飾,同時減弱HOS1對該蛋白的泛素化,以增強ICE蛋白的穩定性,從而通過其下游CBF網絡增強植物的耐凍性[95]。Zhang等研究發現,OsMAPK3磷酸化OsICE1后,OsICE1直接調控OsTPP1的表達,從而促進海藻糖合成,正調控水稻耐寒性[65]。來自蘿卜的脅迫反應基因RsICE1在提高水稻耐冷性方面發揮積極作用,可能是通過與RsICE1基因互作上調OsDREBL和OsTPP1在低溫脅迫下的表達水平[46]。Yang等研究發現,在低溫條件下,MdbHLH4通過直接與MdCBF1和MdCBF3啟動子結合來抑制其表達,從而負調控蘋果耐寒性。它還與MdCICE1L相互作用,并抑制MdICE1L與MdCBF1/MdCBF3啟動子的結合,從而抑制其表達[67]。
3.3 ROS及植物激素介導的信號通路
冷脅迫會引起超氧陰離子自由基(O-2·)、過氧化氫(H2O2)和羥基自由基(·OH)等ROS的過量積累,對細胞中的蛋白質、脂質和核酸具有高反應性和毒性,最終導致細胞死亡。清除過量ROS是應對冷脅迫的必要手段之一。植物進化出高效的酶促和非酶促抗氧化系統來保護自身免受氧化損傷。酶促抗氧化系統包括超氧化物歧化酶(SOD)、過氧化物酶(POD)、過氧化氫酶(CAT)、抗壞血酸過氧化物酶(APX)、谷胱甘肽過氧化物酶(GPX)、脫氫抗壞血酸還原酶(DHAR)、單脫氫抗壞血酸還原酶(MDHAR)、谷胱甘肽還原酶(GR)、谷胱甘肽S-轉移酶(GST)等[96]。在低溫脅迫前后,編碼ROS清除酶基因(NtAPX、NtSOD和NtCAT)的轉錄水平在PubHLH1過表達株系中上調[55]。NtbHLH123過表達植株在冷脅迫下表現出更低的電解質外滲率,降低丙二醛含量,減少H2O2和ROS的積累,這有助于緩解冷脅迫處理后細胞膜的氧化損傷[50]。Geng等研究發現,CsbHLH18介導的耐冷性可能至少部分是通過直接或間接調控抗氧化基因(CsPOD、CsSOD、CsCAT)來調節抗氧化系統[76]。Luo等從多花薔薇(Rosa multiflora)中分離得到RmICE1,在煙草中過表達RmICE1通過調節抗氧化酶系統介導的ROS清除和脅迫響應基因的表達來增強抗寒性[77]。非酶促抗氧化系統包括還原型谷胱甘肽(GSH)、類胡蘿卜素、抗壞血酸(AsA)、類黃酮、花青素、生育酚/維生素E等[96]。低溫條件下,MdbHLH3通過調控花青素合成基因MdDFR和MdU-FGT的表達提高花青素的積累,從而提高蘋果植株的抗寒性[66]。番茄中參與花青素合成調控的AH(Hoffmans Anthocyaninless)基因編碼1個與矮牽牛(Petunia hybrida)AN1和TT8具有高度序列同源性的bHLH蛋白,通過促進低溫環境下花青素的合成,提高植物對低溫的耐受性。此外,與AH過表達株系相比,AH突變體在低溫條件下表現出更多的ROS積累和組成型激活的防御反應,表明AH在抵御低溫脅迫中起關鍵作用[74]。Miura等為鑒定SlICE1的功能,構建過表達番茄株系,同源表達SlICE1基因通過增強冷響應基因SlCBF1和SlDRCi7的表達以及抗壞血酸的積累,以提高番茄的耐冷性[73]。
植物激素茉莉酸(JA)作為脂肪酸代謝的衍生物,廣泛存在于植物中,它可以通過與其他激素或轉錄因子相互作用完成多種生物過程的調控,在調節植物生長、發育和抗逆等方面發揮著重要的作用。JA信號通路核心轉錄因子MYC2在調控植物耐寒中發揮了不可或缺的作用[51]。Zhao等首次報道了關于MYC2與ICE1的相互作用,在香蕉(Musa acuminaa)中,JA信號調控因子MaMYC2s與MaICE1相互作用,MeJA顯著誘導了ICE-CBF冷響應途徑基因MaCBF1、MaCBF2、MaCOR1、MaKIN2、MaRD2和MaRD5的表達,從而增強其抗寒性[85]。Ming等研究發現,由于JA的誘導,PtrMYC2與PtrBADH-1的啟動子結合并激活其表達促進甜菜堿的生物合成,從而提高枳的耐冷性[81]。Min等研究發現,SlMYC2可能是通過改善抗氧化酶系統和增加脯氨酸和番茄紅素水平參與了MeJA介導果實的耐寒性(圖2)[97]。
4 展望
作為真核生物中存在最廣泛的轉錄因子之一,bHLH家族成員眾多,功能豐富。越來越多的證據表明,bHLH轉錄因子在調節植物的低溫脅迫應答反應中起著關鍵作用。bHLH轉錄因子通過特異性結合脅迫相關基因啟動子區的順式作用元件,來調控其轉錄表達,從而調節植物低溫應激反應。本文基于bHLH的結構特征與分類,綜述了其在調控靶基因和參與信號通路中的作用。但目前,關于bHLH轉錄因子在調控植物響應低溫脅迫中的作用研究還不夠深入,特別是在轉錄和蛋白水平。未來可以通過轉錄調控因子與其他因子協同作用,轉錄后和翻譯后修飾等方式研究bHLH轉錄因子的功能。借助高通量測序、全基因組關聯分析和蛋白質組學分析等生物技術,可以了解不同的bHLH相關網絡,解釋植物對低溫脅迫的響應機制。鑒于此,bHLH可以被利用并應用于培育抗逆性提高的作物品種方面。
參考文獻:
[1]Pearce R S. Plant freezing and damage[J]. Annals of Botany,2001,87(4):417-424.
[2]Sun X,Wang Y,Sui N.Transcriptional regulation of bHLH during plant response to stress[J]. Biochemical and Biophysical Research Communications,2018,503(2):397-401.
[3]Hao Y Q,Zong X M,Ren P,et al. Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis[J]. International Journal of Molecular Sciences,2021,22(13):7152.
[4]Jolma A,Yin Y M,Nitta K R,et al. DNA-dependent formation of transcription factor pairs alters their binding specificity[J]. Nature,2015,527:384-388.
[5]Chinnusamy V,Ohta M,Kanrar S,et al. ICE1:a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis[J]. Genes & Development,2003,17(8):1043-1054.
[6]Fursova O V,Pogorelko G V,Tarasov V A. Identification of ICE2,a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana[J]. Gene,2009,429(1/2):98-103.
[7][JP3]Li F,Guo S Y,Zhao Y,et al. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang wild rice confers freezing and salt tolerance in transgenic Arabidopsis[J]. Plant Cell Reports,2010,29(9):977-986.
[8]Feller A,Machemer K,Braun E L,et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. The Plant Journal:for Cell and Molecular Biology,2011,66(1):94-116.
[9]Atchley W R,Terhalle W,Dress A. Positional dependence,cliques,and predictive motifs in the bHLH protein domain[J]. Journal of Molecular Evolution,1999,48(5):501-516.
[10]Nair S K,Burley S K. Recognizing DNA in the library[J]. Nature,2000,404:715-717.
[11]Murre C,McCaw P S,Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding,daughterless,MyoD,and myc proteins[J]. Cell,1989,56(5):777-783.
[12][JP3]Carretero-Paulet L,Galstyan A,Roig-Villanova I,et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis,poplar,rice,moss,and algae[J]. Plant Physiology,2010,153(3):1398-1412.
[13]Ludwig S R,Habera L F,Dellaporta S L,et al. Lc,a member of the maize R gene family responsible for tissue-specific anthocyanin production,encodes a protein similar to transcriptional activators and contains the myc-homology region[J]. Proceedings of the National Academy of Sciences of the United States of America,1989,86(18):7092-7096.
[14]Bailey P C,Martin C,Toledo-Ortiz G,et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana[J]. The Plant Cell,2003,15(11):2497-2502.
[15]Li X X,Duan X P,Jiang H X,et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiology,2006,141(4):1167-1184.
[16]Sun H,Fan H J,Ling H Q.Genome-wide identification and characterization of the bHLH gene family in tomato[J]. BMC Genomics,2015,16(1):9.
[17]Zhou X,Liao Y L,Kim S U,et al. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba[J]. Scientific Reports,2020,10:13723.
[18]Kou X B,Xiong C L,Wang D Q,et al. Comparative analysis of bHLH transcription factors in five Rosaceae species, and expression analysis of PbbHLHs in response to drought stress in pear[J]. Research Square, 2020(9):10.21203/rs.3.rs-78164/v1.
[19]Jaillon O, Aury J M,Noel B,et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla[J]. Nature, 2007, 449(7161): 463-467.
[20]Zhang L,Chen W,Liu R R,et al. Genome-wide characterization and expression analysis of bHLH gene family in physic nut (Jatropha curcas L.)[J]. PeerJ,2022,10:e13786.
[21]Li J J,Li X,Han P,et al. Genome-wide investigation of bHLH genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus L.[J]. International Journal of Biological Macromolecules,2021,189:72-83.
[22]Bano N,Patel P,Chakrabarty D,et al. Genome-wide identification,phylogeny,and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum)[J]. Physiology and Molecular Biology of Plants,2021,27(8):1747-1764.
[23]Ke Q L,Tao W J,Li T T,et al. Genome-wide identification,evolution and expression analysis of basic helix-loop-helix (bHLH) gene family in barley (Hordeum vulgare L.)[J]. Current Genomics,2020,21(8):621-644.
[24]Wang R Q,Zhao P,Kong N N,et al. Genome-wide identification and characterization of the potato bHLH transcription factor family[J]. Genes,2018,9(1):54.
[25]Zhang C H,Feng R C,Ma R J,et al. Genome-wide analysis of basic helix-loop-helix superfamily members in peach[J]. PLoS One,2018,13(4):e0195974.
[26]Yang M,Zhou C P,Yang H,et al. Genome-wide analysis of basic helix-loop-helix transcription factors in papaya (Carica papaya L.)[J]. PeerJ,2020,8:e9319.
[27]Zhang T T,Lv W,Zhang H S,et al. Genome-wide analysis of the basic helix-loop-helix (bHLH) transcription factor family in maize[J]. BMC Plant Biology,2018,18(1):235.
[28]Li J L,Wang T,Han J,et al. Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber[J]. BMC Plant Biology,2020,20(1):272.
[29]Zhang Z S,Chen J,Liang C L,et al. Genome-wide identification and characterization of the bHLH transcription factor family in pepper (Capsicum annuum L.)[J]. Frontiers in Genetics,2020,11:570156.
[30]Dong H Z,Chen Q M,Dai Y Q,et al. Genome-wide identification of PbrbHLH family genes,and expression analysis in response to drought and cold stresses in pear (Pyrus bretschneideri)[J]. BMC Plant Biology,2021,21(1):86.
[31]Liang J X,Fang Y Y,An C,et al. Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress[J]. International Journal of Biological Macromolecules,2023,225:389-403.
[32]Jiang H M,Liu L L,Shan X Z,et al. Genome-wide identification and expression analysis of the bHLH gene family in cauliflower (Brassica oleracea L.)[J]. Physiology and Molecular Biology of Plants,2022,28(9):1737-1751.
[33]Wang R H,Li Y Y,Gao M G,et al. Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L.[J]. BMC Genomics,2022,23(1):548.
[34]Hong Y Q,Ahmad N,Tian Y Y,et al. Genome-wide identification,expression analysis,and subcellular localization of Carthamus tinctorius bHLH transcription factors[J]. International Journal of Molecular Sciences,2019,20(12):3044.
[35]Fan Y,Yang H,Lai D L,et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in sorghum[Sorghum bicolor (L.) Moench][J]. BMC Genomics,2021,22(1):415.
[36]Mao T Y,Liu Y Y,Zhu H H,et al. Genome-wide analyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera[J]. PeerJ,2019,7:e7153.
[37]Gao C,Sun J L,Wang C Q,et al. Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its roles in pod development[J]. PLoS One,2017,12(7):e0181843.
[38]Tian S Y,Li L J,Wei M,et al. Genome-wide analysis of basic helix-loop-helix superfamily members related to anthocyanin biosynthesis in eggplant (Solanum melongena L.)[J]. PeerJ,2019,7:e7768.
[39]Ni L J,Wang Z Q,Fu Z K,et al. Genome-wide analysis of basic helix-loop-helix family genes and expression analysis in response to drought and salt stresses in Hibiscus hamabo Sieb.et Zucc[J]. International Journal of Molecular Sciences,2021,22(16):8748.
[40]Ma P C,Rould M A,Weintraub H,et al. Crystal structure of MyoD bHLH domain-DNA complex:perspectives on DNA recognition and implications for transcriptional activation[J]. Cell,1994,77(3):451-459.
[41]Heim M A,Jakoby M,Werber M,et al. The basic helix-loop-helix transcription factor family in plants:a genome-wide study of protein structure and functional diversity[J]. Molecular Biology and Evolution,2003,20(5):735-747.
[42]Yang X Y,Wang R,Hu Q L,et al. DlICE1,a stress-responsive gene from Dimocarpus longan,enhances cold tolerance in transgenic Arabidopsis[J]. Plant Physiology and Biochemistry,2019,142:490-499.
[43]Wang X P,Song Q P,Guo H,et al. StICE1 enhances plant cold tolerance by directly upregulating StLTI6A expression[J]. Plant Cell Reports,2023,42(1):197-210.
[44]Feng H L,Ma N N,Meng X,et al. A novel tomato MYC-type ICE1-like transcription factor,SlICE1a,confers cold,osmotic and salt tolerance in transgenic tobacco[J]. Plant Physiology and Biochemistry,2013,73:309-320.
[45]Zuo Z F,Kang H G,Park M Y,et al. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis[J]. Plant Science:an International Journal of Experimental Plant Biology,2019,289:110254.
[46]Man L L,Xiang D J,Wang L N,et al. Stress-responsive gene RsICE1 from Raphanus sativus increases cold tolerance in rice[J]. Protoplasma,2017,254(2):945-956.
[47]Zhang Z H,Zhu L,Song A P,et al. Chrysanthemum (Chrysanthemum morifolium) CmICE2 conferred freezing tolerance in Arabidopsis[J]. Plant Physiology and Biochemistry,2020,146:31-41.
[48]Xu W R,Jiao Y T,Li R M,et al. Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis[J]. PLoS One,2014,9(7):e102303.
[49]Zuo Z F,Kang H G,Hong Q C,et al. A novel basic helix-loop-helix transcription factor,ZjICE2 from Zoysia japonica confers abiotic stress tolerance to transgenic plants via activating the DREB/CBF regulon and enhancing ROS scavenging[J]. Plant Molecular Biology,2020,102(4):447-462.
[50]Zhao Q,Xiang X H,Liu D,et al. Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis[J]. Frontiers in Plant Science,2018,9:381.
[51]Wang Y C,Xu H F,Liu W J,et al. Methyl jasmonate enhances apple cold tolerance through the JAZ-MYC2 pathway[J]. Plant Cell,Tissue and Organ Culture (PCTOC),2019,136(1):75-84.
[52]Wang R,Yu M M,Xia J Q,et al. Overexpression of TaMYC2 confers freeze tolerance by ICE-CBF-COR module in Arabidopsis thaliana[J]. Frontiers in Plant Science,2022,13:1042889.
[53]Xu W R,Zhang N B,Jiao Y T,et al. The grapevine basic helix-loop-[JP2]helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis[J]. Molecular Biology Reports,2014,41(8):5329-5342.
[54][JP2]Feng X M,Zhao Q,Zhao L L,et al. The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple[J]. BMC Plant Biology,2012,12:22.
[55]Jin C,Huang X S,Li K Q,et al. Overexpression of a bHLH1 transcription factor of Pyrus ussuriensis confers enhanced cold tolerance and increases expression of stress-responsive genes[J]. Frontiers in Plant Science,2016,7:441.
[56]Yao P F,Sun Z X,Li C L,et al. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis[J]. Plant Physiology and Biochemistry,2018,125:85-94.
[57]Wang Z Y,Zhang Y M,Hu H F,et al. CabHLH79 acts upstream of CaNAC035 to regulate cold stress in pepper[J]. International Journal of Molecular Sciences,2022,23(5):2537.
[58]Lee C M,Thomashow M F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana[J]. Proceedings of the National Academy of Sciences of the United States of America,2012,109(37):15054-15059.
[59]Jiang B C,Shi Y T,Zhang X Y,et al. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America,2017,114(32):E6695-E6702.
[60]He Z Y,Zhao T T,Yin Z P,et al. The phytochrome-interacting transcription factor CsPIF8 contributes to cold tolerance in citrus by regulating superoxide dismutase expression[J]. Plant Science,2020,298:110584.
[61]Wang F,Chen X X,Dong S J,et al. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato[J]. Plant Biotechnology Journal,2020,18(4):1041-1055.
[62]Cordeiro A M,Figueiredo D D,Tepperman J,et al. Rice [JP3]phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B[J]. Biochimica et Biophysica Acta,2016,1859(2):393-404.
[63]He Y N,Li Y P,Cui L X,et al. Phytochrome B negatively affects cold tolerance by regulating OsDREB1 gene expression through phytochrome interacting factor-like protein OsPIL16 in rice[J]. Frontiers in Plant Science,2016,7:1963.
[64]Wang Y J,Zhang Z G,He X J,et al. A rice transcription factor OsbHLH1 is involved in cold stress response[J]. Theoretical and Applied Genetics,2003,107(8):1402-1409.
[65]Zhang Z Y,Li J H,Li F,et al. OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance[J]. Developmental Cell,2017,43(6):731-743.e5.
[66]Xie X B,Li S,Zhang R F,et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples[J]. Plant,Cell & Environment,2012,35(11):1884-1897.
[67]Yang J,Guo X,Mei Q L,et al. MdbHLH4 negatively regulates apple cold tolerance by inhibiting MdCBF1/3 expression and promoting MdCAX3L-2 expression[J]. Plant Physiology,2023,191(1):789-806.
[68]Xu H F,Wang N,Wang Y C,et al. Overexpression of the transcription factor MdbHLH33 increases cold tolerance of transgenic apple callus[J]. Plant Cell,Tissue and Organ Culture,2018,134(1):131-140.
[69]Jiang H F,Shi Y T,Liu J Y,et al. Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize[J]. Nature Plants,2022,8:1176-1190.
[70]Yang T R,Yao S F,Hao L,et al. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway[J]. Plant Cell Reports,2016,35(11):2309-2323.
[71]Badawi M,Reddy Y V,Agharbaoui Z,et al. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes[J]. Plant and Cell Physiology,2008,49(8):1237-1249.
[72]Huang X S,Li K Q,Jin C,et al. ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1[J]. Scientific Reports,2015,5:17620.
[73]Miura K,Shiba H,Ohta M,et al. SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato,Solanum lycopersicum[J]. Plant Biotechnology,2012,29(3):253-260.
[74]Qiu Z K,Wang X X,Gao J C,et al. The tomato Hoffmans anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures[J]. PLoS One,2016,11(3):e0151067.
[75]Zhang T G,Mo J N,Zhou K,et al. Overexpression of Brassica campestris BcICE1 gene increases abiotic stress tolerance in tobacco[J]. Plant Physiology and Biochemistry,2018,132:515-523.
[76]Geng J J,Liu J H. The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene[J]. Journal of Experimental Botany,2018,69(10):2677-2692.
[77]Luo P,Li Z Y,Chen W,et al. Overexpression of RmICE1,a bHLH transcription factor from Rosa multiflora,enhances cold tolerance via modulating ROS levels and activating the expression of stress-responsive genes[J]. Environmental and Experimental Botany,2020,178:104160.
[78]Hu Y F,Zhang H J,Gu B,et al. The transcription factor VaMYC2 from Chinese wild Vitis amurensis enhances cold tolerance of grape (V.vinifera) by up-regulating VaCBF1 and VaP5CS[J]. Plant Physiology and Biochemistry:PPB,2022,192:218-229.
[79]Geng J J,Wei T L,Wang Y,et al. Overexpression of PtrbHLH,a basic helix-loop-helix transcription factor from Poncirus trifoliata,confers enhanced cold tolerance in pummelo (Citrus grandis) by modulation of H2O2 level via regulating a CAT gene[J]. Tree Physiology,2019,39(12):2045-2054.
[80]Huang X S,Zhang Q H,Zhu D X,et al. ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase[J]. Journal of Experimental Botany,2015,66(11):3259-3274.
[81]Ming R H,Zhang Y,Wang Y,et al. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis[J]. The New Phytologist,2021,229(5):2730-2750.
[82]Zhou L,He Y J,Li J,et al. An eggplant SmICE1a gene encoding MYC-type ICE1-like transcription factor enhances freezing tolerance in transgenic Arabidopsis thaliana[J]. Plant Biology,2020,22(3):450-458.
[83]Shen T J,Wen X P,Wen Z,et al. Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.)[J]. Scientia Horticulturae,2021,279:109905.
[84]Gao J,Dou T X,He W D,et al. MaMAPK3-MaICE1-MaPOD P7 pathway,a positive regulator of cold tolerance in banana[J]. BMC Plant Biology,2021,21(1):97.
[85]Zhao M L,Wang J N,Shan W,et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit[J]. Plant,Cell & Environment,2013,36(1):30-51.
[86]Peng P H,Lin C H,Tsai H W,et al. Cold response in Phalaenopsis aphrodite and characterization of PaCBF1 and PaICE1[J]. Plant and Cell Physiology,2014,55(9):1623-1635.
[87]Lin Y Z,Zheng H Q,Zhang Q,et al. Functional profiling of EcaICE1 transcription factor gene from Eucalyptus camaldulensis involved in cold response in tobacco plants[J]. Journal of Plant Biochemistry and Biotechnology,2014,23(2):141-150.
[88]Xiang D J,Man L L,Yin K D,et al. Overexpression of a ItICE1 gene from Isatis tinctoria enhances cold tolerance in rice[J]. Molecular Breeding,2013,32(3):617-628.
[89]Chen Y,Jiang J F,Song A P,et al. Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1 Arabidopsis via miR398[J]. BMC Biology,2013,11:121.
[90]Xu M,Li S J,Liu X F,et al. [JP3]Ternary complex EjbHLH1-EjMYB2-EjAP2-1 retards low temperature-induced flesh lignification in loquat fruit[J]. Plant Physiology and Biochemistry,2019,139:731-737.
[91]位志坤,許自成,賈國濤,等. bHLH轉錄因子家族調控植物非 生物逆境脅迫響應的研究進展[J/OL]. 分子植物育種,2022:1-14(2022-06-15)[2024-07-29]. https://kns.cnki.net/kcms/detail/46.1068.S.20220615.0859.002. html.
[92]Hu Y R,Jiang L Q,Wang F,et al. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis[J]. The Plant Cell,2013,25(8):2907-2924.
[93]李 靜,王 永,楊耀東,等. 植物CBF轉錄因子抗寒作用機制研究進展[J]. 江西農業學報,2014,26(1):59-63.
[94]Dong C H,Agarwal M,Zhang Y Y,et al. The negative regulator of plant cold responses,HOS1,is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1[J]. Proceedings of the National Academy of Sciences of the United States of America,2006,103(21):8281-8286.
[95]Miura K,Jin J B,Lee J,et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis[J]. The Plant Cell,2007,19(4):1403-1414.
[96]柯媛媛,陳 翔,倪芊芊,等. 低溫逆境脅迫下小麥ROS代謝及調控機制研究進展[J]. 大麥與谷類科學,2021,38(1):1-6,21.
[97]Min D D,Li F J,Zhang X H,et al. SlMYC2 involved in methyl jasmonate-induced tomato fruit chilling tolerance[J]. Journal of Agricultural and Food Chemistry,2018,66(12):3110-3117.
基金項目:寧夏自然科學基金(編號:2022AAC03010);寧夏重點研發計劃(編號:2022BBF03004)。
作者簡介:李 珊(2000—),女,湖南衡陽人,碩士,主要從事果樹分子育種相關研究。E-mail:lshanym@163.com。
通信作者:尹 曉,博士,講師,主要從事果樹分子育種教學和相關研究。E-mail:yinxiao90@nxu.edu.cn。

