禾荔特晚熟焦核突變體GLL-1全基因組變異分析
丁峰 李浩然 王金英 彭宏祥 何新華 黃小雄 李平 鐘敏芝 覃燕 張樹偉


摘要:【目的】探究禾荔特晚熟焦核突變體GLL-1的全基因組變異情況,為調控荔枝果實成熟期、解析焦核發生分子機制及選育焦核品種提供理論支持。【方法】通過荔枝種質資源普查發現1個禾荔特晚熟焦核突變體GLL-1,對禾荔和GLL-1開展全基因組重測序(測序深度50×),對比分析GLL-1全基因組變異情況。【結果】與禾荔相比,GLL-1果實明顯較大,品質優良,特晚熟,種子變為焦核,可食率明顯提高。從GLL-1基因組獲得320858674個高質量的Clean reads,定位到荔枝參考基因組的高質量Clean reads數占比為96.07%,正確識別率大于Q20的堿基占比為96.48%,正確識別率大于Q30的堿基占比為91.38%,基因組GC含量為35.28%,覆蓋度(大于1×的堿基占比)為97.62%。檢測到9306084個單核苷酸多態性(SNP)位點和759887個小片段插入和缺失(Indel)位點,共導致12621個基因發生變異,其中發生非同義SNP突變的基因8451個,發生Indel的基因4170個。許多與花色素苷生物合成相關的MYB、bHLH、WD40轉錄因子家族基因及參與ABA信號轉導的重要家族基因(bZIP、WRKY、MAPK及PPR)均發生突變。【結論】MYB、bHLH、WD40、bZIP、WRKY、MAPK及PPR等家族基因的突變可能是導致GLL-1果實特晚熟及焦核發生的一個主要原因,推測其在調控荔枝果實發育和焦核發生中發揮關鍵作用。
關鍵詞: 荔枝;焦核突變體;全基因組重測序;變異分析;SNP;Indel
中圖分類號: S667.103.6? ? ? ? ? ? ? ? ? ? ? ? ? ? ?文獻標志碼: A 文章編號:2095-1191(2021)07-1780-10
Genome-wide variation analysis of the aborted-seeded and
late-maturing mutant GLL-1 from Heli
DING Feng1,2,3, LI Hao-ran1, WANG Jin-ying3, PENG Hong-xiang2, HE Xin-hua3,
HUANG Xiao-xiong1, LI Ping4, ZHONG Min-zhi4, QIN Yan4, ZHANG Shu-wei1,2*
(1Guangxi Academy of Agricultural Sciences/Guangxi Crop Genetic Improvement and Biotechnology Laboratory,Nanning? 530007, China; 2 Horticultural Research Institute, Guangxi Academy of Agricultural Sciences,Nanning? 530007, China; 3 College of Agriculture/State Key Laboratory for Conservation and Utilization of Subtropical
Agro-bioresources, Guangxi University, Nanning? 530004, China; 4Agricultural Technology
Extension Station of Madong,Guiping, Guangxi? 537200, China)
Abstract:【Objective】The present study aimed to study the genome-wide variation situation of Lihe super late-matu-ring aborted-seeded mutant GLL-1, and provide theoretical basis for regulating the ripening period of litchi fruit, analy-zing Molecular mechanisms of aborted-seeds, and breeding of aborted-seeded litchi varieties. 【Method】Through the survey of litchi germplasm resources, Lihe super late-maturing aborted-seeded mutant GLL-1 was found, whole-genome resequencing of cultivars Heli and GLL-1 with 50× depth was conducted, and GLL-1 whole-genome variation situations were compared. 【Result】Compared with Heli, the GLL-1 fruit was obviously larger, with good quality, especially late ripening, seeds became into aborted-seeds, the edible rate was greatly improved. Obtained a total of 320858674 high qua-lity clean reads from GLL-1 genome,of which 96.07% were located in the reference genome of litchi,and the percentage of bases with the correct recognition rate greater than Q20 was 96.48%. The percentage of bases with the correct recognition rate greater than Q30 was 91.38%; the genomic GC content was 35.28%,97.62% of bases with a depth of more than 1× were covered. The results revealed 9306084 single nucleotide polymorphisms(SNPs) and 759887 insertion and deletion of small fragments(Indels),which conferred 12621 mutant genes. A total of 8451 non-synonymous SNP mutations and 4170 Indel genes were identified. It was worth noting that many anthocyanin biosynthesis associated? transcription factor family genes including MYB,bHLH and WD40 and important family genes involving in ABA signal transduction including bZIP,WRKY,MAPK and PPR were mutated. 【Conclusion】Mutations in family genes,including MYB,bHLH,WD40,bZIP,WRKY,MAPK and PPR,may be a major cause of the super late ripening and the aborted-seeded traits of GLL-1 fruit,which play a key role in regulating litchi fruit development and aborted-seeded traits.
Key words: litchi; aborted-seed mutant; whole-genome resequencing; mutation analysis; SNP;Indel
Foundation item:National Natural Science Foundation of China(32060659); Guangxi Natural Science Foundation(2017GXNSFAA198350,2018GXNSFAA050089,2018GXNSFAA294034)
0 引言
【研究意義】荔枝(Litchi chinensis Sonn.)為無患子科(Sapindaceae)荔枝屬(Litchi Sonn.)亞熱帶常綠果樹,起源于我國,因其果實形、色、香、味俱佳和營養豐富而被譽稱為“嶺南果王”“人間仙果”及“佛果”等。目前我國是荔枝的最大生產國,主產區分布在廣東、廣西及海南等地,隨著荔枝產業的快速發展,產業結構問題日益突出,尤其是各主產區品種栽培結構不合理,特早熟和特晚熟品種所占比例很小,而中熟品種所占比例較大,再加上荔枝6、7月份采后保鮮難,導致廣大果農豐產年而不豐收,消減了果農栽培荔枝的積極性,出現了荔枝果園無人管理現象,嚴重影響我國荔枝產業的發展。造成上述結果的主要原因是缺乏品質優良的特早熟和特晚熟荔枝品種。由于荔枝熟期育種缺乏基礎理論指導,導致品種選育進程慢。因此,以禾荔及其特晚熟焦核芽變新種質GLL-1為材料,通過重測序挖掘調控荔枝果實發育速度和焦核發生的基因,為今后荔枝的熟期育種及焦核品種的選育提供基因資源和材料支撐。【前人研究進展】荔枝果皮著色直觀反映了果實成熟進程,其實質是一個花色素苷合成積累的過程,最終成熟時果皮呈現出鮮艷的紅色。在此過程中,MYB、bHLH(Basic helix-loop-helix)和WD40三大類轉錄因子相互作用形成MBW復合體(MYB-bHLH-WD40)共同調控花色素苷的生物合成(Baudry et al.,2004;Zimmermann et al.,2004;Xu et al.,2015)。已有研究證實,MYB轉錄因子在擬南芥、蘿卜等植物花色素苷積累過程中發揮關鍵作用(Borevitz et al.,2000;Zuluaga et al.,2008;Lim et al.,2016)。bHLH轉錄因子在調控植物花色素苷生物合成中也發揮關鍵作用,如擬南芥bHLH轉錄因子突變體tt8、eg3和egl3幼苗和種皮花色素苷積累量明顯減少(Nesi et al.,2000;Zhang et al.,2003);蘋果MdbHLH3和MdbHLH33可與MYB轉錄因子互作,進而調控果實花色素苷的合成(Espley et al.,2007;Xie et al.,2012)。WD40蛋白是調控花色素苷生物合成的另一個重要轉錄因子,如矮牽牛PhAN11是第一個發現的參與花色素苷合成調控的WD40蛋白(de Vetten et al.,1997);擬南芥WD40蛋白ttg1突變體種子的花色素苷合成顯著受抑制(Walker et al.,1999);在果樹中,蘋果MdTTG1、石榴PgWD40、VvWDR1(葡萄)等WD40蛋白均參與其果實花色素苷的生物合成(Brueggemann et al.,2010;Matus et al.,2010;Ben-Simhon et al.,2011)。在荔枝果皮著色研究方面,Lai等(2014,2015,2016)采用轉錄組測序技術分析荔枝果皮成熟過程中基因轉錄本的變化,研究發現有2個LcbHLH轉錄因子能與LcMYB1轉錄因子相互作用調控荔枝果皮花色素苷的積累。由于荔枝種子敗育變為焦核,使得可食率大幅提高,深受廣大消費者的親睞,因此,焦核是評估荔枝果實品質的一個重要指標。研究發現,焦核品種幼胚中的脫落酸(ABA)含量急劇上升改變生長促進物質與生長抑制物質的正常配比,是導致其胚敗育的一個重要原因(周碧燕等,1998;張以順等,2003),表明ABA在荔枝種子敗育發生過程中發揮關鍵作用。【本研究切入點】隨著高通量測序技術的快速發展,不同作物全基因組測序工作相繼完成,目前已對水稻(Hiroki et al.,2013)、菜豆(Jeremy et al.,2014)、大豆(Qi et al.,2014)及番茄(Lin et al.,2014)等作物進行遺傳分析。荔枝全基因組測序也已完成,使荔枝全基因組水平的遺傳分析成為可能。但目前鮮見有關荔枝果實成熟期的調控機制及種子焦核發生機制的研究報道,其主要原因在于缺乏理想的試材。而本研究發現的禾荔特晚熟焦核突變體GLL-1可為荔枝果實成熟期的調控機制及焦核發生機制研究提供重要試材。【擬解決的關鍵問題】以禾荔和GLL-1為試材,通過基因組重測序手段分析禾荔特晚熟焦核突變體GLL-1的全基因組變異情況,為荔枝果實成熟期調控及種子焦核發生機制的研究打下理論基礎。
1 材料與方法
1. 1 試驗材料
供試禾荔母樹(高空壓條苗)及其焦核突變體GLL-1的幼葉采自廣西桂平市麻垌鎮的荔枝果園(東經110°9′,北緯23°8′)。DL2000 DNA Marker、瓊脂糖和植物DNA提取試劑盒購自生工生物工程(上海)股份有限公司。主要儀器設備:紫外可見分光光度計UV5Nano(METTLER TOLEDO,瑞士)、電泳儀1645050(Bio-Rad,美國)、HiSeqTM 2500測序儀(Illumina,美國)。
1. 2 試驗方法
1. 2. 1 GLL-1生物學特性觀測 從GLL-1枝條采集接穗進行高接換種試驗,連續6年(2015─2020年)對GLL-1果實主要性狀進行觀測和評價,并與禾荔果實進行比較。
1. 2. 2 禾荔和GLL-1全基因組重測序 采用植物DNA提取試劑盒分別提取禾荔和GLL-1的幼葉總DNA,具體步驟參照其說明書,并利用紫外分光光度計測定其純度和濃度,1.0%瓊脂糖凝膠電泳檢測其完整性。將檢測合格的總DNA樣品交至深圳華大基因股份有限公司進行全基因組重測序,具體步驟:(1)對DNA進行片段化及純化、末端修復、3?端加A及連接測序接頭;(2)通過1.0%瓊脂糖凝膠電泳進行片段大小的選擇,進行PCR擴增構建測序文庫;(3)利用Illumina HiSeqTM 2500測序儀進行測序,測序深度為50×。從基因組測序數據中過濾去除低質量的reads得到高質量的Clean reads,用于后續的生物信息學分析。
1. 2. 3 GLL-1基因組變異檢測及注釋 分別將禾荔和GLL-1的Clean reads與荔枝參考基因組(http://litchidb.genomics.cn/page/species/index.jsp)進行比對,使用GATK進行單核苷酸多態性(SNP)和小片段插入和缺失(Indel)位點檢測(McKenna et al.,2014),并通過二者比較分析GLL-1的SNP和Indel變異情況。使用SnpEff對GLL-1變異的SNP和Indel位點進行注釋。
1. 2. 4 GLL-1變異基因分析 通過生物信息學方法分析挖掘GLL-1與禾荔間的非同義突變SNP及編碼區(CDS)InDel的基因,再通過BLAST將變異基因與NR、SwissProt、GO、COG及KEGG等數據庫進行比對,從而獲得基因功能注釋(Altschul et al.,1997;Ashburner et al.,2000;Tatusov et al.,2000;Minoru et al.,2004;鄧泱泱等,2006)。
2 結果與分析
2. 1 禾荔和GLL-1生物學特性比較結果
據近6年的表型觀察,發現禾荔和GLL-1的花期均為4月中旬左右,但兩者果實發育進度不同,禾荔果實成熟期在7月中下旬,而GLL-1果實成熟期在8月上中旬,相差約15 d左右(圖1和圖2);GLL-1果實較大,平均單果重24 g左右,而禾荔平均單果重19 g左右;禾荔果皮龜裂片平滑,而GLL-1果皮龜裂片錐尖狀突起(圖3);GLL-1果實焦核率高達92%左右,而禾荔只有4%左右(圖3);GLL-1種子變為焦核后可食率明顯提高,達83%左右,而禾荔種子不變成焦核,可食率僅為73%左右,說明GLL-1為特晚熟優稀芽變荔枝新種質。
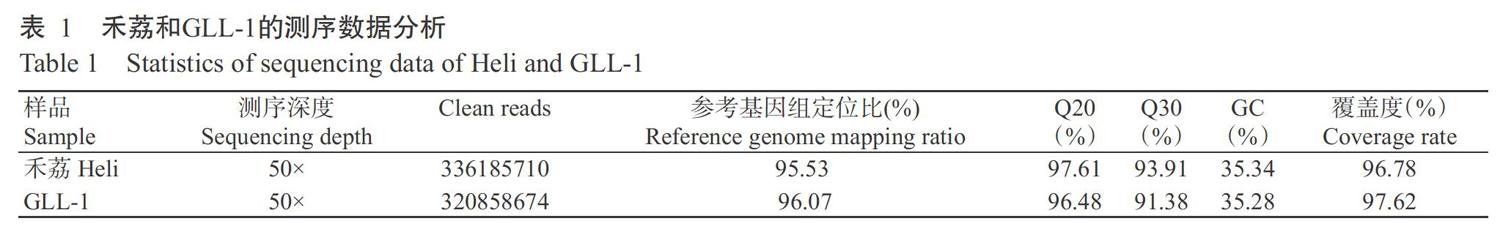
2. 2 禾荔和GLL-1的全基因組重測序結果
利用Illunima HiSeqTM 2500分別對禾荔和GLL-1幼葉DNA進行全基因組重測序。禾荔全基因組重測序結果(表1)顯示,去除帶接頭或低質量的reads后共獲得336185710個Clean reads,定位到荔枝參考基因組的Clean reads占比為95.53%;正確識別率大于Q20的堿基占比97.61%,正確識別率大于Q30的堿基占比為93.91%;基因組GC含量為35.34%,覆蓋度(大于1×的堿基占比)為96.78%。GLL-1基因組重測序結果(表1)顯示,去除帶接頭或低質量的reads后共獲得320858674個Clean reads,定位到荔枝參考基因組的Clean reads占比為96.07%;正確識別率大于Q20的堿基占比為96.48%,正確識別率大于Q30的堿基占比為91.38%;基因組GC含量為35.28%;樣品平均覆蓋深度50×,覆蓋深度大于1×的堿基占比為97.62%。
2. 3 GLL-1基因組變異位點檢測及注釋結果
根據禾荔和GLL-1的Clean reads在荔枝參考基因組進行變異檢測,結果表明GLL-1共發現10065971個變異(Variant)位點,包括9306084個SNP位點和759887個Indel位點;SNP位點中,轉換類型(Transition,Ti)的SNP位點有6684053個,顛換類型(Transversion,Tv)的SNP位點有2622031個;雜合SNP位點有1260043個,純合SNP位點有8046041個,雜合率為15.66%。
使用GATK對GLL-1基因組變異位點進行注釋,結果(表2)顯示,位于CDS序列的SNP位點共計301822個,其中同義突變120761個,非同義突變181061個;位于CDS序列的Indel位點共計8926個,其中包括非3個堿基的整數倍核苷酸缺失突變3204個,3個堿基的整數倍核苷酸缺失突變1779個,非3個堿基的整數倍核苷酸插入突變2353個,3個堿基的整數倍核苷酸插入突變1590個;終止密碼子丟失突變1134個。基于表2注釋信息,經生物信息學分析發現,這些變異共導致12621個基因發生突變,其中發生非同義SNP突變的基因8451個,發生Indel的基因4170個。
2. 4 GLL-1變異基因分析結果
KEGG通路富集分析結果顯示,與禾荔基因組相比,GLL-1有115個植物激素信號傳導相關的基因發生變異。針對GLL-1芽變后成熟期比禾荔明顯延遲(主要體現在果皮花色素苷生物合成變慢)及種子變為焦核不育的特性,對相關候選基因進行挖掘,其中重點挖掘啟動子區域或外顯子區域發生突變的基因,尤其是參與調控花色素苷生物合成的MYB、bHLH和WD40三大類轉錄因子家族基因,結果(表3)發現,GLL-1突變體MYB轉錄因子家族中有15個基因發生突變,其中12個基因啟動子區域發生了SNP突變,2個基因啟動子區域發生了缺失突變,還有一個基因外顯子區域發生了SNP突變;bHLH轉錄因子家族中有11個基因發生突變,其中5個基因啟動子區域發生了SNP突變,2個基因啟動子區域發生了缺失突變,3個基因外顯子區域發生了SNP突變,還有1個基因啟動子區域同時發生了SNP突變和缺失突變;WD40轉錄因子家族中有3個基因發生突變,均為啟動子區域發生了SNP突變。
由表4可知,GLL-1突變體ABA信號轉導通路中15個關鍵基因發生了突變,如2個bZIP轉錄因子基因的啟動子區域發生了SNP突變;5個WRKY轉錄因子基因中,有3個基因的啟動子區域發生了SNP突變,有1個基因的啟動子區域發生缺失突變,有1個基因的啟動子區域發生插入突變;5個MAPK基因中,有4個基因的啟動子區域發生了SNP突變,有1個基因的外顯子區域發生了SNP突變;3個PPR轉錄因子基因中,有2個基因的啟動子區域分別發生了SNP突變及缺失突變,1個基因的外顯子區域發生了SNP突變。以上這些基因的突變可能是造成GLL-1果實相關性狀改變的重要原因,故推測ABA在荔枝種子焦核發生過程中起關鍵作用。
3 討論
本研究通過生物學特性觀察發現,與禾荔相比,其芽變新種質GLL-1果實明顯較大,品質優良,特晚熟,種子變為焦核,可食率明顯提高,同時保存了禾荔豐產、穩產的特性,屬特晚熟優稀荔枝種質資源,適合在晚熟荔枝產區推廣種植,為今后荔枝早、中、晚熟栽培品種結構的優化,延長鮮果產品供應期提供品種支撐,以提高荔枝產業經濟效益。同時,在今后的荔枝雜交育種及研究工作中,GLL-1既可作為父本用于選育特晚熟和焦核優良品種,也可作為研究荔枝果實發育快慢和焦核發生的重要材料。荔枝果皮著色是一個花色素苷生物合成積累的過程,著色快慢代表果實發育的快慢。本研究為了盡量減少測序的假陽性,以50×的荔枝基因組測序深度開展重測序試驗,獲得在啟動子區域和外顯子區域發生突變的MYB、bHLH和WD40轉錄因子基因,其數量分別為15、11和3個,推測MYB、bHLH和WD40三大類轉錄因子在調控花色素苷的生物合成過程中發揮關鍵作用。該結論在其他物種中也得到證實。如轉基因擬南芥中過表達MYB轉錄因子基因AtPAP1可導致其植株變為紫色(Borevitz et al.,2000;Zuluaga et al.,2008);擬南芥bHLH轉錄因子突變體tt8、eg3和egl3的種皮和植株花色素苷積累量均明顯減少(Nesi et al.,2000;Zhang et al.,2003);甜櫻桃MYB轉錄因子PacMYBA可與bHLH轉錄因子相互作用調控花色素苷的合成(Shen et al.,2014);擬南芥WD40蛋白突變體ttg1種子中的花色素苷生物合成受到明顯抑制(Walker et al.,1999;van Nocker and Ludwig,2003;Couture et al.,2006)。此外,本研究發現GLL-1中大量MYB、bHLH和WD40轉錄因子發生突變,突變的位置發生在啟動子區域和外顯子區域。啟動子區域發生突變可能會嚴重影響基因的表達水平,而外顯子區域發生突變可能會嚴重影響到基因的生物學功能,故推測這些基因的突變是造成GLL-1果實晚熟的主要原因,還有待進一步的研究。
與禾荔相比,其芽變新種質GLL-1的重要突變性狀是種子變為焦核,而焦核是評估荔枝果實品質優良的一個關鍵因素。ABA參與調節細胞多種生理過程,包括氣孔關閉、種子發育和萌發等,其在荔枝種子敗育發生過程中起著重要作用(周碧燕等,1998;Finkelstein et al.,2002;張以順等,2003)。此外,研究表明,WRKY18、WRKY40和WRKY60參與ABAR介導的ABA信號轉導途徑,作為轉錄抑制因子互相協作,抑制下游ABA信號調節基因的表達,包括ABI4、ABI5、ABF4和MYB2,進而負調控ABA信號通路(Shang et al.,2010)。PPR蛋白也參與ABA信號轉導過程,包括PPR40(Zsigmond et al.,2008)、ABO5(Liu et al.,2010)、PGN(Laluk et al.,2011)、SLG1(Yuan and Liu,2012)、AHG11(Murayama et al.,2012)、SLO2(Zhu et al.,2014)及SOAR1(Jiang et al.,2015)等。ABA還可誘導ABI5和ABFs等bZIP轉錄因子的表達,其中ABI5是ABA信號轉導的重要正調節子,主要在種子中表達(Finkelstein and Lynch,2000;Lopez-Molina and Chua,2000;Lopez-Molina et al.,2001,2002)。同時,植物MAPK級聯途徑中的相關蛋白通過協同作用參與ABA信號轉導,共同調控植物的生長發育過程(Xing et al.,2008;Jammes et al.,2009)。此外,很多關鍵轉錄因子家族,包括MYC和MYB(Martin and Paz-Ares,1997;Dubos et al.,2010)、bZIP(Jakoby et al.,2002)、WRKY(?lker and Somssich,2004;Rushton et al.,2010)等均需要依賴ABA的逆境信號轉導。因此,本研究主要針對ABA信號轉導通路篩選關鍵突變基因,以期探究GLL-1果實種子焦核突變的分子調控機制。在本研究中,針對ABA信號轉導通路篩選到2個bZIP基因、5個WRKY基因、5個MAPK基因、3個PPR基因、15個MYB基因及11個MYC(bHLH)基因發生了突變,推測MYB、MYC、bZIP、WRKY及PPR等家族基因的突變可能是導致GLL-1種子焦核發生的一個主要原因。基于本研究結果,今后應深入探究荔枝焦核發生的分子調控機制及焦核分子育種技術。
4 結論
MYB、bHLH、WD40、bZIP、WRKY、MAPK及PPR等家族基因的突變可能是導致GLL-1果實特晚熟及種子敗育成焦核的主要原因,推測其在調控荔枝果實發育和焦核發生中發揮關鍵作用。
參考文獻:
鄧泱泱,荔建琦,吳松鋒,朱云平,陳耀文,賀福初. 2006. NR數據庫分析及其本地化[J].計算機工程,32(5):71-73. [Deng Y Y,Li J Q,Wu S F,Zhu Y P,Chen Y W,He F C. 2006. Integrated NR database in protein annotation system and its localization[J]. Computer Engineering,32(5):71-73.]
張以順,向旭,黃上志,傅家瑞. 2003. 荔枝胚敗育過程中內源激素與蛋白質含量的變化[J]. 植物生理與分子生物學學報,29(3):233-238. [Zhang Y S,Xiang X,Huang S Z,Fu J R. 2003. Changes in endogenous hormone and protein content during embryo abortion in litchi[J]. Journal of Plant Physiology and Molecular Biology,29(3):233-238.]
周碧燕,季作梁,葉永昌,招曉東,葉耀雄. 1998. 荔枝果實發育期間內源激素含量的變化[J]. 園藝學報,25(3):236-240. [Zhou B Y,Ji Z L,Ye Y C,Zhao X D,Ye Y X. 1998. Changes of endogenous hormones in litchi fruits during fruit development[J]. Acta Horticulturae Sinica,25(3):236-240.]
Altschul S F,Madden T L,Sch?ffer A A,Zhang J H,Zhang Z,Miller W,Lipman D J. 1997. Gapped BLAST and PS-BLAST:A new generation of protein database search programs[J]. Nucleic Acids Research,25(17):3389-3402. doi:10.1093/nar/25.17.3389.
Ashburner M,Ball C A,Blake J A,Botstein D,Michael-Cherry J. 2000. Gene Ontology:Tool for the unification of biology[J]. Nature Genetics,25(1):25-29. doi:10.1038/75556.
Baudry A,Heim M A,Dubreucq B,Caboche M,Lepiniec L. 2004. TT2,TT8,and TTG specify the synergistically expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana[J]. The Plant Journal,39(3):366-380. doi:10.1111/j.1365-313X.2004.02138.x.
Ben-Simhon Z,Judeinstein S,Nadler-Hassar T,Trainin T,Bar-Yaakov I,Borochov-Neori H,Holland D. 2011. A pomegranate(Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development[J]. Planta,234(5):865-881. doi:10.1007/s00425-011-1438-4.
Borevitz J O,Xia Y,Blount J,Dixon R A,Lamb C. 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis[J]. The Plant Cell,12(12):2383-2394. doi:10.2307/3871236.
Brueggemann J,Weisshaar B,Sagasser M A. 2010. WD40-repeat gene from Malus×domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1[J]. Plant Cell Reports,29(3):285-294. doi:10.1007/s00299-010-0821-0.
Couture J F,Collazo E,Trievel R C. 2006. Molecular recognition of histone H3 by the WD40 protein WDR5[J]. Nature Structural & Molecular Biology,13(8):698-703. doi:10.2210/pdb2h14/pdb.
de Vetten N,Quattrocchio F,Mol J,Koes R. 1997. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast,plants,and animals[J]. Genes & Development,11(11):1422-1434. doi:10.1101/gad.11.11.1422.
Dubos C,Stracke R,Grotewold E,Weisshaar B,Martin C,Lepiniec L. 2010. MYB transcription factors in Arabidopsis[J]. Trends Plant Science,15:573-581. doi:10.1016/j.tplants.2010.06.005.
Espley R V,Hellens R P,Putterill J,Stevenson D E,Kutty-Amma S,Allan A C. 2007. Red colouration in apple fruit is due to the activity of the MYB transcription factor,MdMYB10[J]. The Plant Journal,49(3):414-427. doi:10.1111/j.1365-313X.2006.02964.x.
Finkelstein R R,Gampala S S,Rock C D. 2002. Abscisic acid signaling in seeds and seedlings[J]. The Plant Cell,14(Sl):S15-S45. doi:10.1105/tpc.010441.
Finkelstein R R,Lynch T J. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor[J]. The Plant Cell,12:599-609. doi:10.2307/3871072.
Hiroki T,Akira A,Kentaro Y,Shunichi K,Satoshi N,Chikako M,Aiko U,Hiroe U,Muluneh T,Shohei T,Hideki I,Liliana M C,Sophien K,Ryohei T. 2013. QTL-Seq:Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations[J]. The Plant Journal,74(1):174-183. doi:10.1111/tpj. 12105.
Jakoby M,Weisshaar B,Dr?ge-Laser W,Vicente-Carbajosa J,Tiedemann J,Kroj T,Parcy F,bZIP Research Group. 2002. bZIP transcription factors in Arabidopsis[J]. Trends in Plant Science,7:106-111. doi:10.1016/S1360-1385(01)02223-3.
Jammes F,Song C,Shin D,Munemasa S,Takeda K,Gu D,Cho D,Lee S,Giordo R,Sritubtim S,Leonhardt N,Ellis B E,MurataY,Kwak J M. 2009. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling[J]. Proceedings of the National Academy of Sciences of the United States of America,106(48):20520-20525. doi:10. 1073/pnas.0907205106.
Jeremy S,Mcclean P E,Sujan M,Mamidi S,Wu G A ,Cannon S B,Grimwood J,Enkins J,Shu S Q,Song Q J,Chavarro C,Torres-Torres M,Geffroy V,Moghaddam S M,Gao D Y,Abernathy B,Barry K,Blair M,Brick M A,Chovatia M,Gepts P,Goodstein D M,Gonzales M,Hellsten U,Hyten D L,Jia G F,Kelly J D,Kudrna D,Lee R,Richard M M S,Miklas P N,Osorno J M,Rodrigues J,Thareau V,Urrea C A ,Wang M,Yu Y,Zhang M,Wing R A,Cregan P B,Rokhsar D S,Jackson S A. 2014. A re-ference genome for common bean and genome-wide ana-lysis of dual domestications[J]. Nature Genetics,46:707-713. doi:10.1038/ng.3008.
Jiang S C,Mei C,Liang S,Yu Y T,Lu K,Wu Z,Wang X F,Zhang D P. 2015. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought,salt and cold stresses[J]. Plant Molecular Biology,88:369-385. doi:10.1007/s11103-015-0327-9.
Lai B,Du L,Liu R,Hu B,Su W B,Qin Y H,Zhao J T,Wang H C,Hu G B. 2016. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation[J]. Frontiers in Plant Science,7:166. doi:10.3389/fpls.2016.00166.
Lai B,Hu B,Qin Y,Zhao J T,Wang H C,Hu G B. 2015. Transcriptomic analysis of Litchi chinensis pericarp du-ring maturation with a focus on chlorophyll degradation and flavonoid biosynthesis[J]. BMC Genomics,16:255. doi:10.1186/s12864-015-1433-4.
Lai B,Li XJ,Hu B,Qin Y H,Huang X M,Wang H C,Hu G B. 2014. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes,tissues,develop-mental phases and ABA and light stimuli in Litchi chinensis[J]. PLoS One,9(1): e86293. doi:10.1371/journal.pone.0086293.
Laluk K,Abuqamar S,Mengiste T. 2011. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abio-tic stress tolerance[J]. Plant Physiology,156:2053-2068. doi:10.1104/pp.111.177501.
Lim S,Song J,Kim D,Kim J K,Lee J Y,Kim Y M,Ha S H. 2016. Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1[J]. Plant Cell Reports,35(3):641-653. doi:10. 1007/s00299-015-1909-3.
Lin T,Zhu G,Zhang J,Xu X,Yu Q,Zheng Z,Zhang Z H,Lun Y Y,Li S,Wang X X,Huang Z J,Li J M,Zhang C Z,Wang T T,Zhang Y Y,Wang A X,Zhang Y C,Lin K,Li C Y,Xiong G S,Xue Y B,Mazzucato A,Causse M,Fei Z J,Giovannoni J J,Chetelat R T,Zamir D,Stadler T,Li J F,Ye Z B,Du Y C,Huang S W. 2014. Genomic analyses provide insights into the history of tomato bree-ding[J]. Nature Genetics,46(11):1220-1226. doi:10.1038/ ng.3117.
Liu Y,He J,Chen Z,Ren,X,Hong,X. 2010. ABA overly-sensitive 5(ABO5),encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3,is involved in the abscisic acid response in Arabidopsis[J]. The Plant Journal,63:749-765. doi:10.1111/j. 1365-313X.2010.04280.x.
Lopez-Molina L,Chua N H. 2000. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana[J]. Plant and Cell Physiology,41(5):541-547. doi:10. 1093/pcp/41.5.541.
Lopez-Molina L,Mongrand S,Chua N H. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America,98:4782-4787. doi:10.1073/pnas.081594298.
Lopez-Molina L,Mongrand S,McLachlin,Chait B T,Chua N H. 2002. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination[J]. The Plant Journal,32(3):317-328. doi:10.1046/j.1365-313x. 2002.01430.x.
Martin C,Paz-Ares J. 1997. MYB transcription factors in plants[J]. Trends in Genetic,13:67-73. doi:10.1016/s0168-9525(96)10049-4.
Matus J T,Poupin M J,Canon P,Bordeu E,Alcalde J A,Arce-Johnson P. 2010. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine(Vitis vinifera L.)[J]. Plant Molecular Biology,72(6):607-620. doi:10.1007/s11103-010-9597-4.
McKenna A,Hanna M,Banks E,Sivachenko A,Cibulskis K,Kernytsky A,Garimella K,Altshuler D,Gabriel S,Daly M,Depristo M A. 2014. The genome analysis toolkit:A mapreduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research,20(9):1297-1303. doi:10.1101/gr.107524.110.
Minoru K,Susumu G,Shuichi K,Yasushi O,Masahiro H. 2004. The KEGG resource for deciphering the genome[J]. Nucleic Acids Research,32(22):277-280. doi:10. 1093/nar/gkh063.
Murayama M,Hayashi S,Nishimura N,Ishide M,Kobayashi K,Yagi Y,Asami T,Nakamura T,Shinozaki K,Hirayama T. 2012. Isolation of Arabidopsis ahg11,a weak ABA hypersensitive mutant defective in nad4 RNA editing[J]. Journal of Experimental Botany,63(14):5301-5310. doi:10.1093/jxb/ers188.
Nesi N,Debeaujon I,Jond C,Pelletier G,Caboche M,Lepi-niec L. 2000. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques[J]. The Plant Cell,12(10):1863-1878. doi:10.1105/tpc.12.10.1863.
Qi X,Li M W,Xie M,Liu X,Ni M,Shao G H,Song C,Yim A K Y,Tao Y,Wong F L,Isobe S,Wong C F,Wong K S,Xu C Y,Li C Q,Wang Y,Guan R,Sun F M,Fan G Y,Xiao Z X,Zhou F,Phang T H,Liu X,Tong S W,Chan T F,Yiu S M,Tabata S,Wang J,Xu X,Lam H M. 2014. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing[J]. Nature Communications,5(5):4340. doi:10.1038/ncomms5340.
Rushton P J,Somssich I E,Ringler P,Shen Q J. 2010. WRKY transcription factors[J]. Trends in Plant Science,15(5):247-258. doi:10.1016/j.tplants.2010.02.006.
Shang Y,Yan L,Liu Z Q,Cao Z,Mei C,Xin Q,Wu F Q,Wang X F,Du S Y,Jiang T,Zhang X F,Zhao R,Sun H L,Liu R,Yu Y T,Zhang D P. 2010. The Mg-chelatase H subunit antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition[J]. The Plant Cell,22:1909-1935. doi:10.1105/tpc.110. 073874.
Shen X,Zhao K,Liu L,Zhang K C,Yuan H Z,Liao X,Wang Q,Guo X W,Li F,Li T H .2014. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-co-lored sweet cherry cv. Hong Deng(Prunus avium L.)[J]. Plant and Cell Physiology,55(5):862-880. doi: 10.1093/pcp/pcu013.
Tatusov R L,Galperin M Y,Natale D A,Koonin E V. 2000. The COG database:A tool for genome-scale analysis of protein functions and evolution[J]. Nucleic Acids Resea-rch,28(1):33-36. doi:10.1093/nar/28.1.33.
?lker B,Somssich I E. 2004. WRKY transcription factors: From DNA binding towards biological function[J]. Current Opinion in Plant Biology,7(5):491-498. doi: 10. 1016/j.pbi.2004.07.012.
van Nocker S,Ludwig P. 2003. The WD-repeat protein superfamily in Arabidopsis:Conservation and divergence in structure and function[J]. BMC Genomics,4(1):50. doi:10.1186/1471-2164-4-50.
Walker A R,Davison P A,Bolognesi-Winfield A C,James C M,Srinivasan N,Blundell T L,Esch J J,Marks M D,Gray J C. 1999. The TRANSPARENT TESTA GLABRA1 locus,which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis,encodes a WD40 repeat protein[J]. The Plant Cell,11(7):1337-1350. doi:10.1105/tpc.11.7.1337.
Xie X B,Li S,Zhang R F,Zhao J,Chen Y C,Zhao Q,Yao Y X,You C X,Zhang X S,Hao Y J. 2012. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low tempera-ture in apples[J]. Plant Cell and Environment,35(11):1884-1897. doi:10.1111/j.1365-3040.2012.02523.x.
Xing Y,Jia W,Zhang J. 2008. AtMKK1 mediates ABA induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis[J]. The Plant Journal,54(3):440-451. doi:10.1111/j.1365-313X.2008.034 33.x.
Xu W,Dubos C,Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends in Plant Science,20(3):176-185. doi:10.1016/ j.tplants.2014.12.001.
Yuan H,Liu D. 2012. Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing,plant development,and responses to abiotic stresses in Arabidopsis[J]. The Plant Journal,70:432-444. doi:10. 1111/j.1365-313X.2011.04883.x.
Zhang F,Gonzalez A,Zhao M,Payne C T,Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis[J]. Development,130(20):4859-4869. doi:10.1242/dev.00681.
Zhu Q,Dugardeyn J,Zhang C,Muhlenbock P,Eastmond P J,Valcke R,De C B,Oden S,Karampelias M,Cammue B P A,Prinsen E,Van D S D. 2014. The Arabidopsis tha-liana RNA editing factor SLO2,which affects the mitochondrial electron transport chain,participates in multiple stress and hormone responses[J]. Molecular Plant,7:290-310. doi:10.1093/mp/sst102.
Zimmermann I M,Heim M A,Weisshaar B,Uhrig J F. 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins[J]. The Plant Journal,40(1):22-34. doi:10.1111/j.1365-313X.2004.02183.x.
Zsigmond L,Rigó G,Szarka A,Székely G,Otv?s K,Darula Z,Medzihradszky K F,Koncz C,Koncz Z,Szabados L. 2008. Arabidopsis PPR40 connects abiotic stress respon-ses to mitochondrial electron transport[J]. Plant Physio-logy,146:1721-1737. doi:10.1104/pp.107.111260.
Zuluaga D L,Gonzali S,Loreti E,Pucciariello C,Degl'Innocenti E,Guidi L,Alpi A,Perata P. 2008. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants[J]. Functional Plant Biology,35(7):606-618. doi:10.1071/FP08021.
(責任編輯 陳 燕)

