黃花蒿中青蒿素生物合成相關(guān)轉(zhuǎn)錄因子研究進展
李 琦,高曉悅,陳萬生,譚何新
黃花蒿中青蒿素生物合成相關(guān)轉(zhuǎn)錄因子研究進展
李 琦,高曉悅,陳萬生,譚何新*
海軍軍醫(yī)大學藥學院,上海 200433
以青蒿素為基礎(chǔ)的聯(lián)合療法是瘧疾的首選治療方案,而藥用植物黃花蒿是青蒿素的唯一天然來源,也是目前最主要的青蒿素來源,因此培育高產(chǎn)青蒿素的黃花蒿一直是國際研究熱點。青蒿素是黃花蒿的次生代謝產(chǎn)物,在植物的次生代謝物合成過程中,關(guān)鍵的轉(zhuǎn)錄因子可以調(diào)節(jié)代謝途徑中某個或多個基因的表達,從而調(diào)節(jié)代謝流的方向和速度,決定著代謝物的產(chǎn)量,因此關(guān)鍵轉(zhuǎn)錄因子的表達對于青蒿素的合成非常重要,通過干預(yù)轉(zhuǎn)錄因子的表達也是提高青蒿素產(chǎn)量的重要手段。綜述了黃花蒿中已研究的轉(zhuǎn)錄因子功能及調(diào)控機制,特別是對篩選獲得轉(zhuǎn)錄因子基因的方法進行總結(jié),以期為揭示青蒿素合成調(diào)控網(wǎng)絡(luò)奠定基礎(chǔ)。
黃花蒿;青蒿素;轉(zhuǎn)錄因子;代謝調(diào)控;次生代謝物
據(jù)世界衛(wèi)生組織(World Health Organization,WHO)報道,2018年全球共有2.28億瘧疾病例報告,其中40.5萬例死亡,2010~2018年瘧疾的發(fā)病率及死亡率均呈下降趨勢,特別是重災(zāi)區(qū)非洲的瘧疾死亡率下降尤為明顯,這很大程度上歸功于以青蒿素為基礎(chǔ)的聯(lián)合療法(artemisinin-based combination therapy,ACT)的應(yīng)用,目前ACT仍為WHO推薦的瘧疾首選治療方案[1]。如何高效獲得青蒿素一直是國際研究熱點,化學合成方面,早在1983年瑞士科學家Schmid和Hofheinz就提出了青蒿素的化學全合成方案,但反應(yīng)步驟多、條件苛刻、試劑昂貴、得率低,后續(xù)提出的化學合成方案均存在類似問題[2]。合成生物學方面,Keasling團隊在青蒿酸生物合成方面的研究堪稱合成生物學的典范,然而由于青蒿素的最終合成需要特殊的油性氧化環(huán)境,在酵母中難以實現(xiàn),因此目前較為成熟的方法仍為生物合成青蒿酸[3]。Paddon等[4]提出以青蒿酸為原料的化學半合成方案,最終青蒿素的得率為40%~45%,且存在分離純化困難的問題。目前最為經(jīng)濟的獲得青蒿素的方法仍為從植物中提取,而青蒿素在植株中的含量較低,僅占干質(zhì)量的0.1%~1%[5],隨著青蒿素臨床治療應(yīng)用的開發(fā),其市場需求會進一步加大,因此需要提高青蒿素的產(chǎn)量。
青蒿素是來源于藥用植物黃花蒿L.的次生代謝產(chǎn)物,其生物合成途徑屬于類異戊二烯途徑(圖1),由質(zhì)體中的異戊二烯(meth-ylerythritol phosphate,MEP)途徑提供1個異戊烯基焦磷酸(isopentenyl phosphate,IPP)和細胞質(zhì)中的甲羥戊酸(mevalonic acid,MVA)途徑提供1個二甲基烯丙基焦磷酸(dimethylallyl diphosphate,DMAPP)和IPP,然后1份DMAPP和2份IPP在法尼基焦磷酸合成酶(farnesyl diphosphate synthase,F(xiàn)PS)的催化下聚合生成法尼基焦磷酸(farnesyl diphosphate,F(xiàn)PP)[6-7]。從FPP開始進入青蒿素合成特異的下游代謝途徑,F(xiàn)PP經(jīng)紫穗槐二烯合成酶(amorpha-4,11-diene synthase,ADS)催化生成紫穗槐二烯[8];紫穗槐二烯經(jīng)過3步由細胞色素P450單氧化酶(Cytochrome P450 monooxygenase,CYP71AV1)催化的反應(yīng),分別形成青蒿醇,青蒿醛和青蒿酸[9-11]。青蒿醛可在青蒿醛雙鍵還原酶[artemisinic aldehyde delta-11(13)reductase,DBR2][12]和醛脫氫酶1(aldehyde dehydrogenase 1,ALDH1)[13]的催化下先轉(zhuǎn)化成二氫青蒿醛(dihydroartemisinic aldehyde),然后生成青蒿素的直接前體二氫青蒿酸(dihydroartemisinic acid,DHAA)。同時,青蒿醛也會在CYP71AV1 和 ALDH1的催化下生成青蒿酸(artemisinic acid)[11, 13];從DHAA到青蒿素(artemisinin),以及青蒿酸到青蒿素B(arteannuin B)的轉(zhuǎn)化目前認為是非酶促的光氧化反應(yīng)[14-17]。在植物的次生代謝過程中,轉(zhuǎn)錄因子可以調(diào)節(jié)代謝途徑中的一系列基因,對轉(zhuǎn)錄因子的干預(yù)是一種有效的調(diào)控植物次生代謝產(chǎn)物的方法[18]。本文按照獲得轉(zhuǎn)錄因子基因的不同途徑進行分類,綜述了黃花蒿中轉(zhuǎn)錄因子的研究進展。
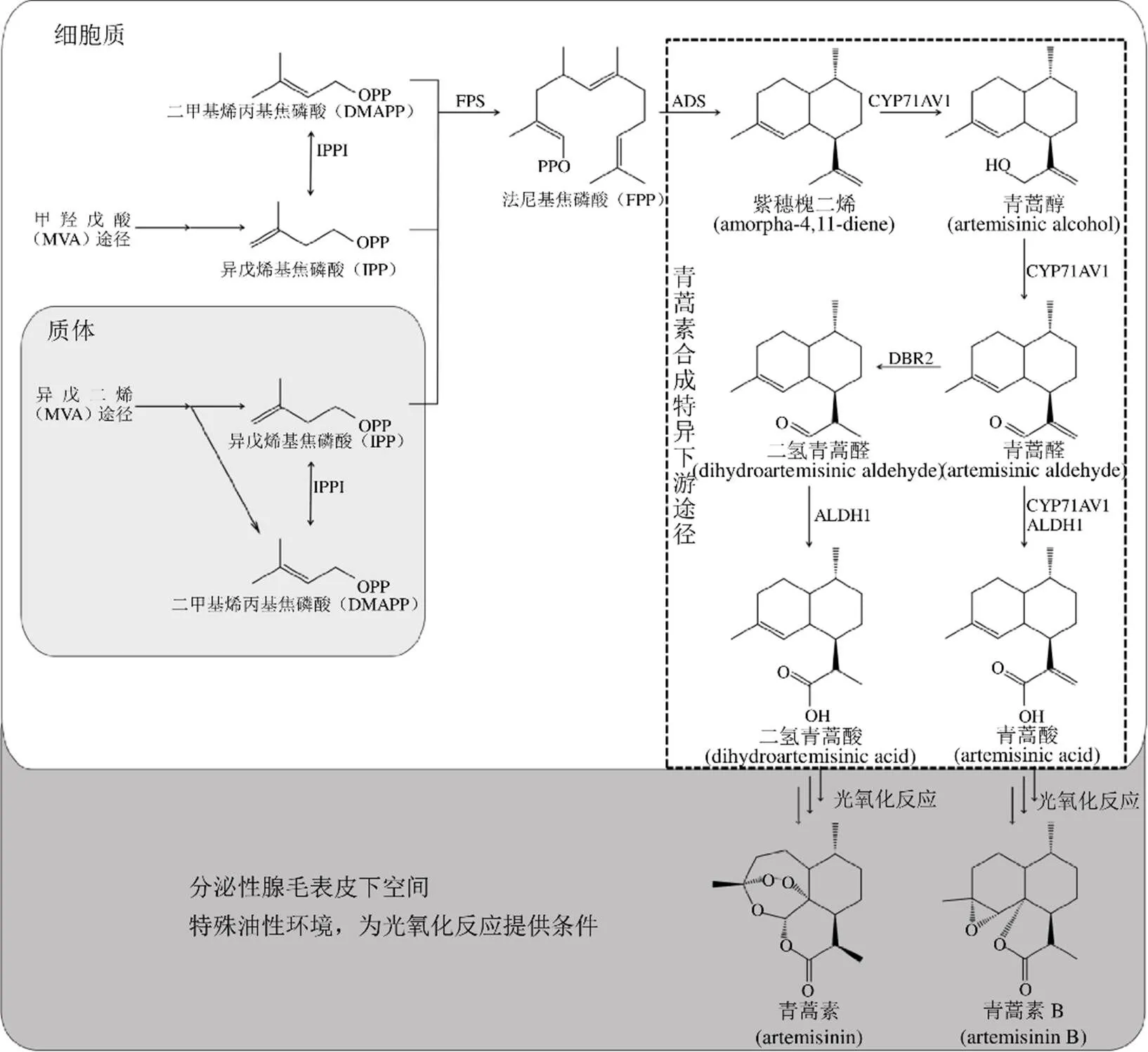
圖1 青蒿素生物合成途徑
1 激素響應(yīng)轉(zhuǎn)錄因子
1.1 脫落酸(abscisic acid,ABA)對黃花蒿bZip1基因(AabZip1)的影響
ABA的處理能提高青蒿素積累[19],在模式植物擬南芥中A類bZip家族轉(zhuǎn)錄因子響應(yīng)ABA信號[20]。Zhang等[21]通過bZip家族保守結(jié)構(gòu)域查詢黃花蒿公共cDNA數(shù)據(jù)庫,得到145個候選基因,其中64個能在分泌性腺毛中表達,通過與擬南芥A類bZip轉(zhuǎn)錄因子基因構(gòu)建系統(tǒng)進化樹,篩選得到6個黃花蒿A類bZip轉(zhuǎn)錄因子基因,再通過在煙草中進行雙熒光素酶實驗驗證能否激活及的表達,獲得目的基因。后續(xù)實驗表明受ABA、干旱、高鹽脅迫的誘導(dǎo)表達,AabZip1通過激活和的表達影響青蒿素的合成,其過表達可使青蒿素和二氫青蒿酸含量分別提高40%~60%、41%~53%。
1.2 茉莉酸(jasmonicacid,JA)和茉莉酸甲酯(methyl jasmonate,MeJA)
1.2.1 黃花蒿基因() bHLH家族的MYC2類轉(zhuǎn)錄因子被認為是參與JA信號通路的核心因子[22-23]。Shen等[24]在黃花蒿葉片cDNA文庫中發(fā)現(xiàn)5個MYC2類轉(zhuǎn)錄因子片段序列,其中1條與MeJA處理后的擬南芥中的MYC2基因的表達模式相同,用RACE方法獲得其全長序列,命名為。后續(xù)研究表明過表達能提高和的轉(zhuǎn)錄水平,可使青蒿素和二氫青蒿酸含量分別提高23%~55%、17%~217%。
1.2.2 黃花蒿基因() JA能促進黃花蒿分泌性腺毛起始[25],JAZ蛋白是茉莉酸信號通路的重要調(diào)控因子,在擬南芥中能影響腺毛的起始[26],Yan等[27]用AaJAZ8篩選黃花蒿幼葉cDNA文庫,發(fā)現(xiàn)1個HDZip家族轉(zhuǎn)錄因子AaHD1能與AaJAZ8相互作用,且受茉莉酸的正調(diào)控。后續(xù)研究表明過表達能在不影響植物生長的情況下增加成熟葉片表面分泌性腺毛密度,使青蒿素含量提高50%。
1.2.3 黃花蒿基因(、) AP2/ERF家族轉(zhuǎn)錄因子參與植物激素信號響應(yīng)及次生代謝[28]。Yu等[29]通過在分泌性腺毛cDNA文庫中查找AP2/ERF家族轉(zhuǎn)錄因子保守結(jié)構(gòu)域,獲得7個片段序列,其中2個符合在花中表達、且受MeJA誘導(dǎo)的表達模式,用RACE的方法獲得這2條序列的全長,即和。后續(xù)研究表明AaERF1、AaERF2均能激活的表達,過表達可使青蒿素和青蒿酸的含量提高19%~67%、11%~76%,過表達為24%~51%、17%~121%,且基因可能通過JA和乙烯信號通路,激活部分防御基因,提高黃花蒿對灰霉病菌Pers.的抗性[30]。
1.3 水楊酸(Salicylicacid,SA)對黃花蒿TGA6基因(AaTGA6)
SA在植物對病原菌的防御響應(yīng)中起重要作用[31],在擬南芥的SA信號通路中TGA2、NPR1是重要調(diào)節(jié)因子[32-33]。Lv等[34]分析黃花蒿轉(zhuǎn)錄組數(shù)據(jù),發(fā)現(xiàn)有6條TGA類轉(zhuǎn)錄因子序列,通過與擬南芥TGA轉(zhuǎn)錄因子基因構(gòu)建系統(tǒng)進化樹,同源性最高的為。后續(xù)研究表明的表達能被AaNPR1增強,被AaTGA3抑制,同時AaTGA6能調(diào)控基因的表達,過表達可使青蒿素含量提高90%~120%。
2 通過分析啟動激活元件獲得轉(zhuǎn)錄因子
2.1 黃花蒿WRKY1基因(AaWRKY1)
Han等[35]分析基因的啟動子發(fā)現(xiàn)其含有2個W-box,可能受WRKY家族轉(zhuǎn)錄因子調(diào)控,通過比對分泌性腺毛的cDNA文庫,發(fā)現(xiàn)其中有1條轉(zhuǎn)錄因子的片段序列,用RACE方法獲得全長序列,命名為。后續(xù)研究表明AaWRKY1能提高及其它青蒿素合成途徑基因的表達,且在腺毛中特異的過表達比在植物中廣泛的過表達能更有效的提高青蒿素的積累。
2.2 黃花蒿MYB1基因(AaMYB1)
Matías-Hernández等[36]發(fā)現(xiàn)多數(shù)青蒿素合成途徑基因啟動子區(qū)域均含有MYB家族轉(zhuǎn)錄因子結(jié)合位點,推測有MYB家族轉(zhuǎn)錄因子參與調(diào)控青蒿素生物合成。分析公共黃花蒿分泌性腺毛cDNA文庫,發(fā)現(xiàn)1個符合R2R3-MYB轉(zhuǎn)錄因子特征的片段,用RACE的方法獲得全長序列命名為。后續(xù)研究表明,的過表達可以提高青蒿素合成途徑基因的表達水平,增加分泌性腺毛數(shù)量及密度,提高青蒿素的積累。
2.3 黃花蒿bHLH1基因(AabHLH1)
Ji等[37]發(fā)現(xiàn)基因啟動子區(qū)域均含有bHLH家族轉(zhuǎn)錄因子結(jié)合的E-box,推測可能被bHLH家族轉(zhuǎn)錄因子調(diào)控,在黃花蒿分泌性腺毛cDNA數(shù)據(jù)庫中分析發(fā)現(xiàn)3條可能的bHLH家族轉(zhuǎn)錄因子基因片段,用RACE方法能獲得了其中2條基因的全長,酵母單雜交實驗及EMSA實驗證明AabHLH1能與E-box結(jié)合,因此選定為目的基因。后續(xù)研究表明,AabHLH1能結(jié)合的啟動子并提高青蒿素合成途徑基因的轉(zhuǎn)錄水平。
2.4 黃花蒿HD8基因(AaHD8)
Yan等[38]分析的啟動子發(fā)現(xiàn)含有L1-box,預(yù)測可能被HD-Zip家族轉(zhuǎn)錄因子結(jié)合,利用酵母單雜交、雙熒光素酶實驗篩選已獲得的HD-Zip IV亞家族轉(zhuǎn)錄因子基因,獲得。后續(xù)研究表明AaHD8能激活的表達,從而起始分泌性腺毛的發(fā)育,另外AaHD8還能與AaMIXTA1相互作用,調(diào)節(jié)腺毛發(fā)育及角質(zhì)層形成。
3 與已知功能基因同源的黃花蒿轉(zhuǎn)錄因子基因
3.1 擬南芥
3.1.1 黃花蒿基因() 擬南芥ORA59能在植物防御中整合JA和乙烯信號[39]。Lu等[40]通過擬南芥基因序列在黃花蒿分泌性腺毛cDNA文庫中查詢,得到1個同源性最高的片段,用RACE方法獲得其全長序列,即。后續(xù)研究表明ABA處理、創(chuàng)傷、寒冷能顯著提高的轉(zhuǎn)錄水平,MeJA、乙烯處理也能輕微的誘導(dǎo)表達,從而參與植物抗逆。
3.1.2 黃花蒿基因() MYB家族轉(zhuǎn)錄因子中MIXTA或MIXTA樣轉(zhuǎn)錄因子能控制多種植物的細胞形態(tài)、腺毛發(fā)育和起始等,如擬南芥AtMYB16和AtMYB106調(diào)控腺毛分枝及表皮細胞形態(tài)[41]。Shi等[42]以擬南芥已知MYB家族轉(zhuǎn)錄因子序列在黃花蒿各組織的轉(zhuǎn)錄組數(shù)據(jù)庫中查詢,獲得的序列與擬南芥及其他物種已知的MIXTA或MIXTA樣轉(zhuǎn)錄因子共同構(gòu)建系統(tǒng)進化樹,篩選獲得與擬南芥、同源性最高的序列,即。后續(xù)研究表明AaMIXTA1可以調(diào)控分泌性腺毛的數(shù)量和角質(zhì)層的生物合成,過表達可以在不影響分泌性腺毛結(jié)構(gòu)的情況下提高青蒿素的積累。
3.1.3 黃花蒿基因() 青蒿素的生物合成量在低溫處理后顯著升高[43-44],而擬南芥中2個低溫相關(guān)的轉(zhuǎn)錄因子AtICE1和ATICE2均屬于bHLH家族[45-46]。Xiang等[47]以bHLH保守結(jié)構(gòu)域在青蒿基因組數(shù)據(jù)庫中查詢,得到基因組中205個結(jié)果、轉(zhuǎn)錄組中122個,按bHLH家族轉(zhuǎn)錄因子特征進行分類,其中屬于與青蒿素合成相關(guān)的V家族轉(zhuǎn)錄因子15個,與擬南芥bHLH轉(zhuǎn)錄因子共同構(gòu)建系統(tǒng)進化樹,2條序列與擬南芥、同源性最高,其中僅受低溫誘導(dǎo)。后續(xù)研究表明,AabHLH112能調(diào)控的表達,從而提高青蒿素合成途徑基因的轉(zhuǎn)錄水平,過表達的株系中青蒿素及二氫青蒿酸含量均顯著上升。
3.2 其他
3.2.1 黃花蒿基因() 長春花的過表達可以增加萜類化合物的積累[48],Lu等[49]以長春花序列作為查詢序列在黃花蒿葉片cDNA文庫中比對,獲得一條片段序列,用RACE方法獲得全長序列,即。后續(xù)研究表明在分泌型腺毛中特異性表達,對有正調(diào)控作用,過表達青蒿素及二氫青蒿酸含量分別上升40%~53%和22%~35%,同時能提高黃花蒿對灰霉病菌的抗性。
3.2.2 黃花蒿基因() 薄荷與萜類合成相關(guān)[50],Kayani等[51]以YABBY家族轉(zhuǎn)錄因子保守結(jié)構(gòu)域在黃花蒿基因組數(shù)據(jù)庫中查詢,將獲得的序列與構(gòu)建系統(tǒng)進化樹,同源性最高的序列即。后續(xù)研究表明能受MeJA的誘導(dǎo),直接結(jié)合、的啟動子,提高青蒿素合成途徑基因當?shù)霓D(zhuǎn)錄水平,過表達可以使青蒿素及二氫青蒿酸的含量升高。
4 轉(zhuǎn)錄組、基因組數(shù)據(jù)分析
4.1 黃花蒿TAR1基因(AaTAR1)
Graham等[52]對黃花蒿的遺傳圖譜進行分析的文章中,預(yù)測了7個可能與腺毛發(fā)育相關(guān)的基因,Tan等[53]通過分析黃花蒿轉(zhuǎn)錄組數(shù)據(jù)及同源比對,篩選出其中的1個AP2/ERF家族轉(zhuǎn)錄因子,命名為AaTAR1。后續(xù)研究表明AaTAR1可以控制蠟質(zhì)、角質(zhì)物質(zhì)的合成,能直接影響分泌型腺毛的形態(tài)發(fā)育,并通過結(jié)合青蒿素合成路徑上的基因啟動子,來控制青蒿素的合成。過表達能提高青蒿素合成途徑基因的轉(zhuǎn)錄水平,并提高黃花蒿葉片和花蕾中青蒿素、青蒿酸及二氫青蒿酸的含量。
4.2 黃花蒿SPL2基因(AaSPL2)
在模式植物擬南芥中,有研究表明SPL轉(zhuǎn)錄因子參與調(diào)控植物的次生代謝[54-55]。Lv等[56]以SPL家族轉(zhuǎn)錄因子保守結(jié)構(gòu)域在黃花蒿轉(zhuǎn)錄組數(shù)據(jù)庫中查詢,得到14個結(jié)果,其中含有MicroRNA156識別位點的9條;用JA處理黃花蒿,檢測9條SPL轉(zhuǎn)錄因子基因的表達水平發(fā)現(xiàn)和響應(yīng)了JA信號,其中的表達模式與青蒿素合成途徑基因的表達模式更為接近,因此選擇作為目的基因。后續(xù)研究表明,基因可激活的轉(zhuǎn)錄,從而介導(dǎo)JA對青蒿素合成的調(diào)控,過表達能提高青蒿素合成途徑基因的轉(zhuǎn)錄水平,使青蒿素和二氫青蒿酸的含量提高33%~86%和26%~159%。
4.3 黃花蒿GSW1基因(AaGSW1)
多條WRKY家族轉(zhuǎn)錄因子已被證實參與調(diào)控青蒿素的生物合成,但它們均非腺毛特異表達轉(zhuǎn)錄因子,Chen等[57]以WRKY保守結(jié)構(gòu)域序列在青蒿各組織轉(zhuǎn)錄組數(shù)據(jù)庫中比對,共獲得122條序列,其中42條能在腺毛中表達,通過與腺毛特異表達基因,如、等,在各組織中的表達模式進行比較,獲得表達模式最為接近的1條,命名為。后續(xù)研究表明AaGSW1是腺毛特異表達轉(zhuǎn)錄因子,受JA信號途徑中的AaMYC2和ABA信號途徑的AabZip1調(diào)控,過表達能提高及的轉(zhuǎn)錄水平,使青蒿素含量提高39%~43%。
4.4 黃花蒿NAC1基因(AaNAC1)
NAC家族轉(zhuǎn)錄因子在提高植物對灰霉病菌抗性,及調(diào)控萜類物質(zhì)合成方面發(fā)揮作用[58-59]。Lv等[60]分析分泌性腺毛轉(zhuǎn)錄組數(shù)據(jù)庫,選出RPKM(Reads Per Kilobase per Million mapped reads)值最高的10條NAC家族轉(zhuǎn)錄因子基因序列,其中能受SA和MeJA的誘導(dǎo),作為目的基因進行研究。后續(xù)研究發(fā)現(xiàn)能受脫水、低溫、SA、MeJA的誘導(dǎo),過表達能使青蒿素和二氫青蒿酸的含量升高,提高黃花蒿對干旱、灰霉病菌的抗性。
5 展望
青蒿素是中醫(yī)藥對世界的重大貢獻,隨著新適應(yīng)癥的開發(fā),青蒿素的需求量會進一步增大,解析青蒿素的合成路徑及調(diào)控網(wǎng)絡(luò),并利用分子育種的手段獲得高青蒿素含量的黃花蒿意義重大。青蒿素的產(chǎn)量與分泌性腺毛的發(fā)育情況密切相關(guān),同時黃花蒿也被認為是研究分泌性腺毛發(fā)育的潛在模式植物,而轉(zhuǎn)錄因子無論在青蒿素的生物合成途徑中,還是在腺毛的發(fā)育過程中均具有關(guān)鍵的調(diào)控作用,因此轉(zhuǎn)錄因子是研究的重要靶標。目前對黃花蒿轉(zhuǎn)錄因子的研究雖然已經(jīng)涉及到AP2/ERF、WRKY、MYB、bZip、bHLH等多個家族,但植物轉(zhuǎn)錄因子數(shù)量龐大,調(diào)控網(wǎng)絡(luò)復(fù)雜,當前對黃花蒿轉(zhuǎn)錄因子調(diào)控作用的認識仍相對有限。隨著研究技術(shù)手段的發(fā)展,特別是轉(zhuǎn)錄組、基因組等組學技術(shù)及單細胞測序等最新技術(shù)開始應(yīng)用在黃花蒿的研究中,將更有利于發(fā)掘重要的功能基因,完善對青蒿素生物合成、分泌性腺毛發(fā)育等重要科學問題的認識。
利益沖突 所有作者均聲明不存在利益沖突
[1] World Health Organization.2019 [M]. Geneva: WHO, 2019.
[2] Schmid G, Hofheinz W. Total synthesis of qinghaosu [J]., 1983, 105: 624-625.
[3] Ro D K, Paradise E M, Ouellet M,. Production of the antimalarial drug precursor artemisinic acid in engineered yeast [J]., 2006, 440(7086): 940-943.
[4] Paddon C J, Westfall P J, Pitera D J,. High-level semi-synthetic production of the potent antimalarial artemisinin [J]., 2013, 496(7446): 528-532.
[5] Mutabingwa T K. Artemisinin-based combination therapies (ACTs): Best hope for malaria treatment but inaccessible to the needy! [J]., 2005, 95(3): 305-315.
[6] Schramek N, Wang H, R?misch-Margl W,. Artemisinin biosynthesis in growing plants of. A13CO2study [J]., 2010, 71(2/3): 179-187.
[7] Ma D M, Li G, Alejos-Gonzalez F,. Overexpression of a type-I isopentenyl pyrophosphate isomerase ofin the cytosol leads to high arteannuin B production and artemisinin increase [J]., 2017, 91(3): 466-479.
[8] Wallaart T E, Bouwmeester H J, Hille J,.-4,11-diene synthase: Cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin [J]., 2001, 212(3): 460-465.
[9] Wang H Z, Han J L, Kanagarajan S,. Trichome-specific expression of the-4,11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in, as reported by a promoter-GUS fusion [J]., 2013, 81(1/2): 119-138.
[10] Wang Y, Yang K, Jing F,. Cloning and characterization of trichome-specific promoter of cpr71av1 gene involved in artemisinin biosynthesis inL. [J]., 2011, 45(5): 817-824.
[11] Teoh K H, Polichuk D R, Reed D W,.L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin [J]., 2006, 580(5): 1411-1416.
[12] Zhang Y S, Teoh K H, Reed D W,. The molecular cloning of artemisinic aldehyde Delta11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in[J]., 2008, 283(31): 21501-21508.
[13] Teoh K, Polichuk D, Reed D,. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in[J]., 2009, 87: 635-642.
[14] Lommen W J, Elzinga S, Verstappen F W,. Artemisinin and sesquiterpene precursors in dead and green leaves ofL. crops [J]., 2007, 73(10): 1133-1139.
[15] Lommen W J, Schenk E, Bouwmeester H J,. Trichome dynamics and artemisinin accumulation during development and senescence ofleaves [J]., 2006, 72(4): 336-345.
[16] Brown G D, Sy L K.transformations of dihydroartemisinic acid inplants [J]., 2004, 60(5): 1139-1159.
[17] Brown G D, Sy L K.transformations of artemisinic acid inplants [J]., 2007, 63(38): 9548-9566.
[18] Verpoorte R, Memelink J. Engineering secondary metabolite production in plants [J]., 2002, 13(2): 181-187.
[19] Jing F Y, Zhang L, Li M Y,. Abscisic acid (ABA) treatment increases artemisinin content inby enhancing the expression of genes in artemisinin biosynthetic pathway [J]., 2009, 64(2): 319-323.
[20] Jakoby M, Weisshaar B, Dr?ge-Laser W,. bZIP transcription factors in[J]., 2002, 7(3): 106-111.
[21] Zhang F, Fu X, Lv Z,. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in[J]., 2015, 8(1): 163-175.
[22] Dombrecht B, Xue G P, Sprague S J,. MYC2differentially modulates diverse jasmonate-dependent functions in[J]., 2007, 19(7): 2225-2245.
[23] Zhang H B, Bokowiec M T, Rushton P J,. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis [J]., 2012, 5(1): 73-84.
[24] Shen Q, Lu X, Yan T X,. The jasmonate-responsive AaMYC2transcription factor positively regulates artemisinin biosynthesis in[J]., 2016, 210(4): 1269-1281.
[25] Maes L, van Nieuwerburgh F C, Zhang Y S,. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin inplants [J]., 2011, 189(1): 176-189.
[26] Qi T C, Song S S, Ren Q C,. The Jasmonate- ZIM-domain proteins interact with the WD-Repeat/ bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in[J]., 2011, 23(5): 1795-1814.
[27] Yan T X, Chen M H, Shen Q,. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in[J]., 2017, 213(3): 1145-1155.
[28] Brown R L, Kazan K, McGrath K C,. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of[J]., 2003, 132(2): 1020-1032.
[29] Yu Z X, Li J X, Yang C Q,. The jasmonate- responsive AP2/ERF transcription factors AaERF1and AaERF2positively regulate artemisinin biosynthesis inL [J]., 2012, 5(2): 353-365.
[30] Lu X, Jiang W M, Zhang L,. AaERF1positively regulates the resistance toin[J]., 2013, 8(2): e57657.
[31] Raskin I, Skubatz H, Tang W,. Salicylic acid levels in thermogenic and non-thermogenic plants [J]., 1990, 66(4): 369-373.
[32] Fan W H, Dong X N.interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in[J]., 2002, 14(6): 1377-1389.
[33] Rochon A, Boyle P, Wignes T,. The coactivator function ofNPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines [J]., 2006, 18(12): 3670-3685.
[34] Lv Z, Guo Z, Zhang L,. Interaction of bZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in[J]., 2019, 70(15): 3969-3979.
[35] Han J L, Wang H Z, Lundgren A,. Effects of overexpression of AaWRKY1on artemisinin biosynthesis in transgenicplants [J]., 2014, 102: 89-96.
[36] Matías-Hernández L, Jiang W M, Yang K,. AaMYB1 and its orthologue AtMYB61 affect terpene metabolism and trichome development inand[J]., 2017, 90(3): 520-534.
[37] Ji Y P, Xiao J W, Shen Y L,. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in[J]., 2014, 55(9): 1592-1604.
[38] Yan T, Li L, Xie L,. A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in[J]., 2018, 218(2): 567-578.
[39] Pré M, Atallah M, Champion A,. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense [J]., 2008, 147(3): 1347-1357.
[40] Lu X, Jiang W, Zhang L,. Characterization of a novel ERF transcription factor inand its induction kinetics after hormones and stress treatments [J]., 2012, 39(10): 9521-9527.
[41] Oshima Y, Shikata M, Koyama T,. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development inand[J]., 2013, 25(5): 1609-1624.
[42] Shi P, Fu X Q, Shen Q,. The roles of AaMIXTA1 in regulating the initiation of glandular trichomes and cuticle biosynthesis in[J]., 2018, 217(1): 261-276.
[43] Liu W, Wang H, Chen Y,. Cold stress improves the production of artemisinin depending on the increase in endogenous jasmonate [J]., 2017, 64(3): 305-314.
[44] Wallaart T E, Pras N, Beekman A C,. Seasonal variation of artemisinin and its biosynthetic precursors in plants ofof different geographical origin: Proof for the existence of chemotypes [J]., 2000, 66(1): 57-62.
[45] Chinnusamy V, Ohta M, Kanrar S,. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in[J]., 2003, 17(8): 1043-1054.
[46] Fursova O V, Pogorelko G V, Tarasov V A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in[J]., 2009, 429(1/2): 98-103.
[47] Xiang L E, Jian D Q, Zhang F Y,. The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1in[J]., 2019, 70(18): 4835-4848.
[48] van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism [J]., 2000, 289(5477): 295-297.
[49] Lu X, Zhang L, Zhang F Y,. AaORA, a trichome-specific AP2/ERF transcription factor of, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to[J]., 2013, 198(4): 1191-1202.
[50] Wang Q, Reddy V A, Panicker D,. Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5from spearmint () [J]., 2016, 14(7): 1619-1632.
[51] Kayani S I, Shen Q, Ma Y N,. The YABBY family transcription factor AaYABBY5directly targets cytochrome P450 monooxygenase (CYP71AV1) and double-bond reductase 2 (DBR2) involved in artemisinin biosynthesis in[J]., 2019, 10: 1084.
[52] Graham I A, Besser K, Blumer S,. The genetic map ofL. identifies loci affecting yield of the antimalarial drug artemisinin [J]., 2010, 327(5963): 328-331.
[53] Tan H X, Xiao L, Gao S H,. TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in[J]., 2015, 8(9): 1396-1411.
[54] Gou J Y, Felippes F F, Liu C J,. Negative regulation of anthocyanin biosynthesis inby a miR156-targeted SPL transcription factor [J]., 2011, 23(4): 1512-1522.
[55] Yu Z X, Wang L J, Zhao B,. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli () by the miR156-targeted SPL transcription factors [J]., 2015, 8(1): 98-110.
[56] Lv Z, Wang Y, Liu Y,. The SPB-box transcription factorpositively regulates artemisinin biosynthesis inL. [J]., 2019, 10: 409.
[57] Chen M H, Yan T X, Shen Q,. GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in[J]., 2017, 214(1): 304-316.
[58] Bu Q, Jiang H, Li C B,. Role of theNAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses [J]., 2008, 18(7): 756-767.
[59] Nieuwenhuizen N J, Chen X, Wang M Y,. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors [J]., 2015, 167(4): 1243-1258.
[60] Lv Z, Wang S, Zhang F Y,. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought andin[J]., 2016, 57(9): 1961-1971.
Research on transcription factors related to artemisinin biosynthesis in
LI Qi, GAO Xiao-yue, ZHANG Lei, CHEN Wan-sheng, TAN He-xin
School of Pharmacy, Navy Medical University, Shanghai 200433, China
Artemisinin-based combination therapy is the preferred treatment for malaria. The medicinal plantis the only natural source and the main source of artemisinin. It is a hotspot to cultivate a strain ofwith high artemisinin content. Artemisinin is a secondary metabolite of. In the process of plant secondary metabolism, transcription factors play important roles in regulating a series of genes in the metabolic pathway, thereby regulating the direction and speed of metabolic flow. Therefore, the intervention of transcription factor by genetic engineering is an important method to regulate plant secondary metabolism. This article summarizes the functions and regulatory mechanism of transcription factors had been studied in. In particular, the methods for screening these transcription factors genes were summarized in order to provide a reference for the finding of key functional genes.
L.; artemisinin; transcription factor; metabolic regulation; secondary metabolite
R282.12
A
0253 - 2670(2021)06 - 1827 - 08
10.7501/j.issn.0253-2670.2021.06.032
2020-06-03
國家自然科學基金資助項目(81673529);科技部新藥創(chuàng)制重大專項(2017ZX09101002-003-002)
李 琦(1992—),男,碩士研究生,研究方向為中藥材品質(zhì)改良。E-mail: liqi0121@163.com
譚何新,女,副教授,博士,碩士生導(dǎo)師,主要從事中藥資源研究。Tel/Fax: (021)81871370 E-mail: hexintan@163.com
[責任編輯 時圣明]

