pyIFPNI: A package for querying and downloading plant fossil data from the IFPNI
Bailong Zhao
a State Key Laboratory of Plant Diversity and Specialty Crops,Institute of Botany,Chinese Academy of Sciences,Beijing,100093,China b University of Chinese Academy of Sciences,Beijing,100049,China
The International Fossil Plant Names Index(IFPNI,2014-onwards)not only serves as an online gateway to the fossil plant name registry for the global scientific community,but also serves as a comprehensive and dynamic archive of fossil plants (Doweld,2016,2022).Since its establishment in 2014,the IFPNI has accumulated more than 80,000 entries of fossil plant names,along with information on over 10,000 documents and over 6000 paleobotanists,all supported by exhaustive data,demonstrating the scope of the IFPNI's work(IFPNI,2014-onwards).As it can meticulously trace the fossil plant name record,the IFPNI is an indispensable resource for both paleobotanists and systematic botanists,providing invaluable insights and information.However,unlike other palaeobotanical databases(i.e.,The Palaeobotanical Database (PBDB) and the Geobiodiversity Database(GBDB);Fan et al.,2011,2013;Deng et al.,2020),the IFPNI lacks a download function.The storage of information from the IFPNI requires manual data for each record,leading to a long and tedious process.Therefore,a crucial task is to create efficient methods to preserve the data obtained from the IFPNI.
Python is a highly popular programming language known for its versatility,user-friendliness,and widespread adoption.In this project,I have developed pyIFPNI,an open-source Python package that simplifies the process of retrieving and acquiring fossil plant name records from IFPNI.This innovative tool integrates a wide range of search techniques available on IFPNI,enabling smooth conversion of the obtained results into a structured pandas dataframe format,which can be conveniently saved as CSV files.Consequently,this streamlined process not only accelerates data collection and preservation but also ensures that subsequent data analysis tasks are efficiently executed,thereby guaranteeing the overall efficiency of the entire workflow.
Researchers have the flexibility to employ different query methods while accessing fossil plant name records.These methods enable them to efficiently navigate through the records by utilizing various classification levels.This functionality is achieved through the implementation of specific methods,which are explained as follows (Fig.1):
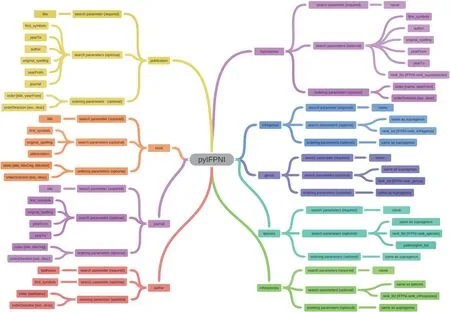
Fig.1.Schematic representation of the methods in the pyIFPNI package.The overview illustrates the complete framework of the pyIFPNI package and presents a detailed breakdown of the specific parameters for each method.
Supragenus: Thesupragenus() function enables researchers to query fossilized plant name records at taxonomic ranks higher than the genus level.This functionality proves invaluable when searching for plant names associated with specific supragenus categories.
Genus: Thegenus() function assists in querying fossilized plant name records at the genus level.
Infragenus: To further refine the search below the genus level,theinfragenus() function comes into play.
Species: When researchers focus on the most fundamental taxonomic rank,thespecies() function becomes relevant.They can utilize this method to query fossilized plant name records specifically associated with particular species or genera.
Infraspecies: Theinfraspecies() function allows researchers to delve even deeper into the taxonomic hierarchy,exploring fossilized plant name records below the species level.This method is particularly useful for investigating subspecies,varieties,or forms of fossilized plants.
When utilizing thesupragenus(),genus(),infragenus(),species(),andinfraspecies() methods in pyIFPNI,users have the flexibility to include additional parameters for more specific queries.These parameters include the following:
Author: Users can specify the author parameter to search for fossil plant name records attributed to specific authors.
Rank List: By specifying the rank parameter,users can narrow down their search to specific taxonomic ranks within the supragenus,genus,infragenus,species,or infraspecies methods.
Original Spelling:Users have the option to include the“original spelling” parameter when searching for fossil plant names.This parameter allows users to find names based on their original spellings,which can be helpful for historical or variant name inquiries.
Publication Year Range: Researchers have the flexibility to define a publication year range parameter while conducting searches for fossil plant name records.This feature enables them to retrieve names that were published within a specific time frame,allowing for more targeted and focused research.
Paleoregion: The “paleoregion” parameter can also be utilized by users when using thespecies() andinfraspecies() methods.This parameter allows researchers to apply a filter to their search,narrowing down the results to specific paleoregions of interest.
The methodspublication(),book(),andjournal() are commonly used in bibliographic databases to query specific types of sources.These methods allow users to retrieve references based on the type of publication they belong to.Here's a brief explanation of each method:
publication(): This method retrieves all types of publications,including books,journal articles,conference proceedings,reports,theses,etc.When using thepublication()method,you generally get a broader range of results encompassing various publication types.
book():This method specifically retrieves references that belong to books.It is useful when searching for information contained within published books.Using thebook() method helps narrow down your search to relevant book titles and their associated details such as author(s),title,publisher,year,etc.
journal(): This method focuses on retrieving references from academic journals.If you are specifically interested in scholarly articles published in journals,using thejournal() method can help filter out other types of publications.
Within the pyIFPNI library,there is a specialized method calledauthor()that empowers users to explore paleobotanists listed in the IFPNI database.This dedicated functionality facilitates convenient access to information regarding specific authors who have made significant contributions to the field of paleobotany,with their work thoroughly documented in the IFPNI repository.By making use of theauthor() method,researchers can efficiently retrieve relevant data related to these paleobotanists and gain valuable insights into their notable contributions.
These advanced search methods encompass exploring higher taxonomic groups related to the target taxon,investigating lower taxonomic groups associated with the target taxon,searching for taxa published in the target publication,and querying for taxa and publications attributed to the targeted paleobotanist.There are 15 advanced methods provided by pyIFPNI.Detailed information and related examples of these methods can be found at https://github.com/WDragon101/pyIFPNI.
The primary objective of pyIFPNI is to retrieve records of fossil plant names through query methods based on taxonomic hierarchy.Each method operates within a specific taxonomic search range,which can be divided into two groups.The first group,includingsupragenus(),genus(),andinfragenus(),explicitly targets names above the species level (excluding the species level),resulting in a limited number of outcomes.The second group consists ofspecies()andinfraspecies()methods,intended to search for names at and below the species level,potentially yielding more outcomes than the previous group.In this section,I will briefly introduce the usage and examples of two fundamental functions,supragenus() andspecies(),which are more commonly used.The remaining functions,including both basic and advanced,are documented at https://github.com/WDragon101/pyIFPNI.
Let us proceed with the following assumption: Our targeted taxonomic group is Berberidaceae,which unequivocally surpasses the level of genus.Hence,we shall employ thesupragenus()method to facilitate our analysis.To ensure the utmost precision in retrieving name records,we will establish a search range spanning from 1753 to 2023,comprehensively encompassing the entirety of recorded instances from the earliest documented occurrence to the present day.This objective can be achieved by explicitly specifying the parameters yearFrom as 1753 and yearTo as 2023 or by omitting these parameters altogether.
Another crucial parameter,rank_list,plays a pivotal role in further refining the scope of our search.In this specific example,the rank_list parameter encompasses the values [“Family”,“Order”],thereby restricting the search solely to these particular taxonomic ranks.Naturally,if required,rank_list can accommodate multiple taxonomic ranks.To determine the permissible values for the rank_list parameter in thesupragenus() method,you may utilizeIFPNI.rank_supragenus.keys().Subsequently,select the desired combination of ranks accordingly(Fig.S1).
The process of searching for species names through the use of thespecies() method parallels thesupragenus() method.The practical application of thespecies() includes the exploration of all names of a genus,which is not achievable through the methods of the first group.For instance,thespecies()method can be applied to extract all species names in the genusBerberisin combination with the same remaining parameters as utilized in thesupragenus()method.By doing so,you can efficiently obtain the entire catalog of fossil species that are related toBerberis(Fig.S2).
To use the pyIFPNI package in R,you can follow these steps.
1.Install the reticulate package: install.packages("reticulate")
2.Load the reticulate package:library(reticulate)
3.Create a Python environment: use_python("
4.Install the pyIFPNI package:py_install("pyIFPNI")
5.Import the pyIFPNI module: pyIFPNI <-import("pyIFPNI")
6.The functions provided by pyIFPNI can be accessed using thepyIFPNI$function_name() syntax.For example,thesupragenus()function can be used by callingpyIFPNI$supragenus().
One drawback is that there is currently no equivalent R package available for pyIFPNI.As a temporary solution,the aforementioned method can be used to invoke pyIFPNI in R.However,recognizing the widespread use of R in the scientific community,I am committed to developing an R version and will soon release relevant updates on https://github.com/WDragon101/pyIFPNI.
I developed pyIFPNI,a sophisticated Python library designed to enable users to explore and acquire fossil plant names from the esteemed International Fossil Plant Names Index.This comprehensive library not only facilitates diverse taxonomic searches but also provides advanced search criteria comparable to those found on IFPNI's official website.A notable feature of this library is its ability to seamlessly integrate search results into a structured dataframe format.This functionality streamlines data analysis and empowers researchers to conveniently store and utilize their findings in the future.To further enhance its utility for paleobotanists and botanists,I aim to bolster the search engine capabilities of pyIFPNI.This entails implementing cutting-edge depth search and taxon search algorithms,which will yield more exhaustive and precise outcomes.These refinements are intended to enhance the overall accessibility and functionality of the library,catering to the needs of the scientific community more effectively.
Author contributions
B.Z.developed the idea of the software,produced the package and write all Python codes,and wrote the paper.
Declaration of competing interest
The author declares that I have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
I thank the anonymous reviewers for their helpful comments.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.12.001.
- 植物多樣性的其它文章
- Intraspecific floral colour variation in three Pedicularis species
- Photosynthetic response dynamics in the invasive species Tithonia diversifolia and two co-occurring native shrub species under fluctuating light conditions
- Reproductive height determines the loss of clonal grasses with nitrogen enrichment in a temperate grassland
- Sex-specific facilitation and reproduction of the gynodioecious cushion plant Arenaria polytrichoides on the Himalaya-Hengduan mountains,SW China
- Different mechanisms underlie similar species-area relationships in two tropical archipelagoes
- Genetic analyses of ancient tea trees provide insights into the breeding history and dissemination of Chinese Assam tea (Camellia sinensis var. assamica)

