Syntheses, Crystal Structures and DNA-Binding Properties of Co/CdComplexes with Quinoline Thiosemicarbazone Ligand
MAO Pan-DongYAN Ling-Ling*,WANG Wen-JingYANG Qian-QianCUI Meng-QiangWANG Yuan*,WU Wei-Na(Department of Physics and Chemistry, Henan Polytechnic University, Jiaozuo, Henan 454000, China)(School of Materials Science and Engineering, Henan Polytechnic University, Jiaozuo, Henan 454000, China)
Syntheses, Crystal Structures and DNA-Binding Properties of Co/CdComplexes with Quinoline Thiosemicarbazone Ligand
MAO Pan-Dong1YAN Ling-Ling*,1WANG Wen-Jing2YANG Qian-Qian2CUI Meng-Qiang1WANG Yuan*,1WU Wei-Na1
(1Department of Physics and Chemistry, Henan Polytechnic University, Jiaozuo, Henan 454000, China)
(2School of Materials Science and Engineering, Henan Polytechnic University, Jiaozuo, Henan 454000, China)
Two complexes [CoL2]Cl·2CH3OH (1) and [CdL2] (2) (HL=(quinolin-8-ylmethylene)thiosemicarbazide) have been synthesized and structurally determined by single-crystal X-ray diffraction. The results show that the metal ion in each complex with a distorted octahedron geometry is surrounded by two anionic thiosemicarbazone ligand with N2S donor set. Complexes 1 and 2 can bind to DNA via electrostatic intercalation and partial intercalation modes, respectively. CCDC: 1436092, 1; 1436093, 2.
quinoline; thiosemicarbazone; complex; crystal structure; DNA-biding property
Thiosemicarbazones (TSCs) and their metal complexes, especially the transition metal, have attracted intensity attention in the coordination chemistry because of their high biological and pharmaceutical activities, such as antibacterial, antiviral, antifungal, and antitumor activity[1]. In fact, the mechanism of antitumor action is still controversial in many respects and has been identified including ribonucleotide reductase inhibition, metal dependent radical damage, DNA binding and inhibition of protein synthesis[1-10]. Up to now, a large amount of TSCs metals containing six-member heterocycles have been found to possess considerable antitumor activity[2,4-10]. However, the studies on the complexes with TSCs bearing condensed heterocycles are relatively few[3,11-13]. Recently, several Cucomplexes with quinolin-8-ylmethylene) thiosemicarbazide have been reported to be potential antitumor agents[12-13]. In this paper, the structures andDNA-binding properties of its Co/Cdcomplexes have been discussed in detail.
1 Experimental
1.1Materials and measurements
Solvents and starting materials for syntheses were purchased commercially and used as received. Elemental analyses were carried out on an Elemental Vario EL analyzer. The IR spectra (ν=4 000~400 cm-1) were determined by the KBr pressed disc method on a Bruker V70 FT-IR spectrophotometer. DNA-Binding Properties of both complexes are measured using literature method via absorption and emission spectra[14]. The UV spectra were recorded on a Purkinje General TU-1800 spectrophotometer. Fluorescence spectra were determined on a Varian CARY Eclipse spectrophotometer, in the measurements of emission and excitation spectra the pass width is 5 nm.
1.2Preparations of the ligand and complexes 1 and 2
The TSC ligand HL (Scheme 1) was produced according to the literature method[13]. The complexes 1 and 2 were generated by reaction of HL (5mmol) with equimolar of CoCl2or Cd(NO3)2in CH3OH (10 mL) solution, respectively. Crystals of 1 and 2 suitable for X-ray diffraction analysis were obtained by evaporating the corresponding reaction solutions at room temperature.
1: purple block. Anal. Calcd. For C24H26N8O2S2ClCo (%): C 46.72; H 4.25; N 18.16. Found (%): C 46.62; H 4.44; N 18.03. FT-IR (cm-1):ν(C=N) 1 590, ν(N=C)quinoline1 482,ν(C-S) 768.
2: yellow plate. Anal. Calcd. for C22H18N8S2Cd(%): C 46.28; H 3.18; N 19.63. Found (%): C 46.44; H 3.29; N 19.48. FT-IR (cm-1):ν(C=N) 1 591,ν(N=C)quinoline1 437,ν(C-S) 730.
1.3X-ray crystallography
The X-ray diffraction measurement for complexes 1 (size: 0.10 mm×0.08 mm×0.08 mm) and 2 (size: 0.15 mm×0.14 mm×0.06 mm) was performed on a Bruker SMART APEXⅡCCD diffractometer equipped with a graphite monochromatized Mo Kα radiation (λ= 0.071 073 nm) by using φ-ω scan mode. Semi-empirical absorption correction was applied to the intensity data using the SADABS program[15]. The structures were solved by direct methods and refined by full matrix least-square on F2using the SHELXTL-97 program[16]. All non-hydrogen atoms were refined anisotropically. All H atoms were positioned geometrically and refined using a riding model. Details of the crystal parameters, data collection and refinements for complexes 1 and 2 are summarized in Table 1.
CCDC: 1436092, 1; 1436093, 2.
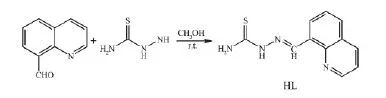
Scheme 1 Synthetic route for HL
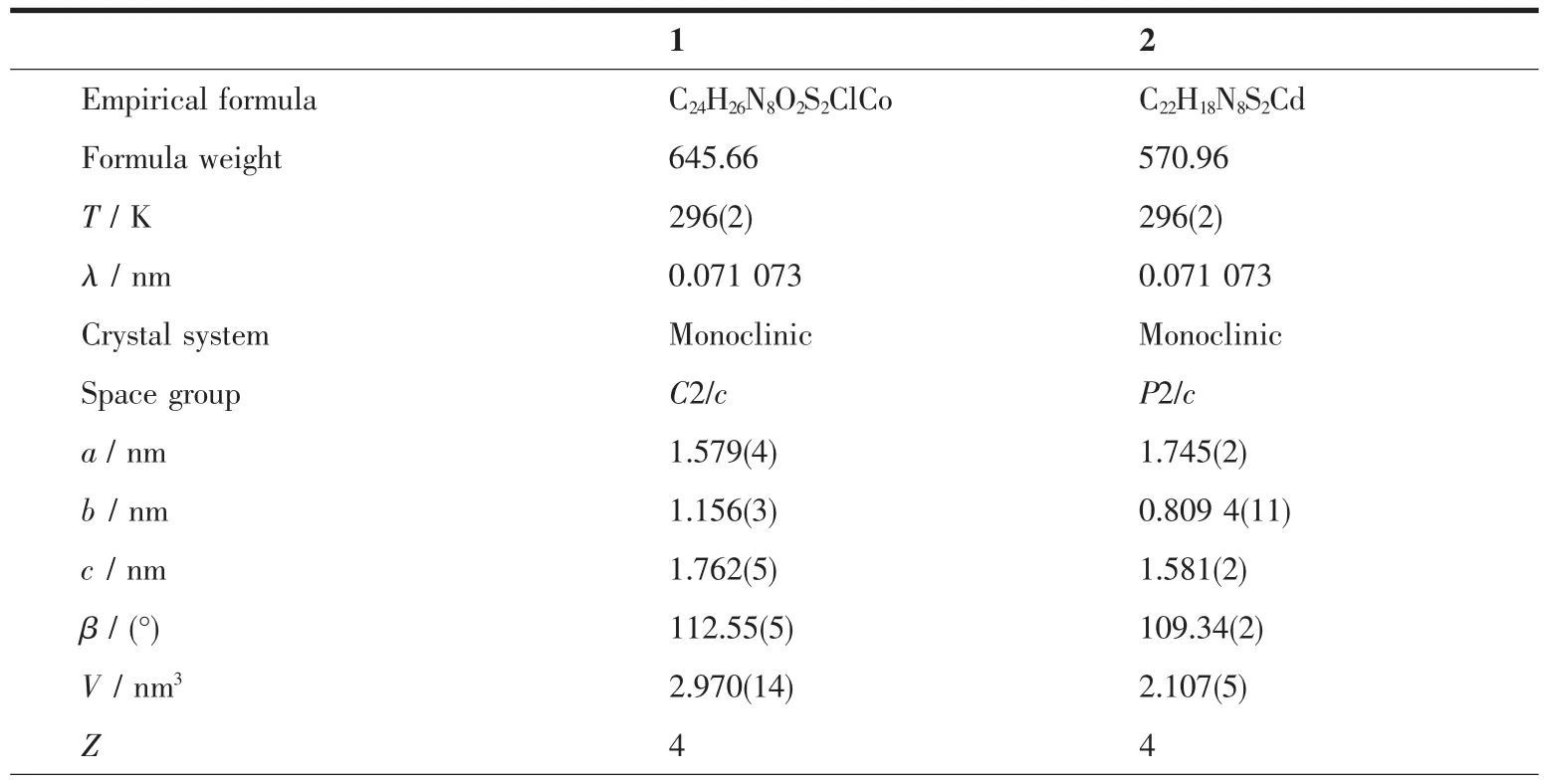
Table 1 Crystal data and structure refinement for complexes 1 and 2
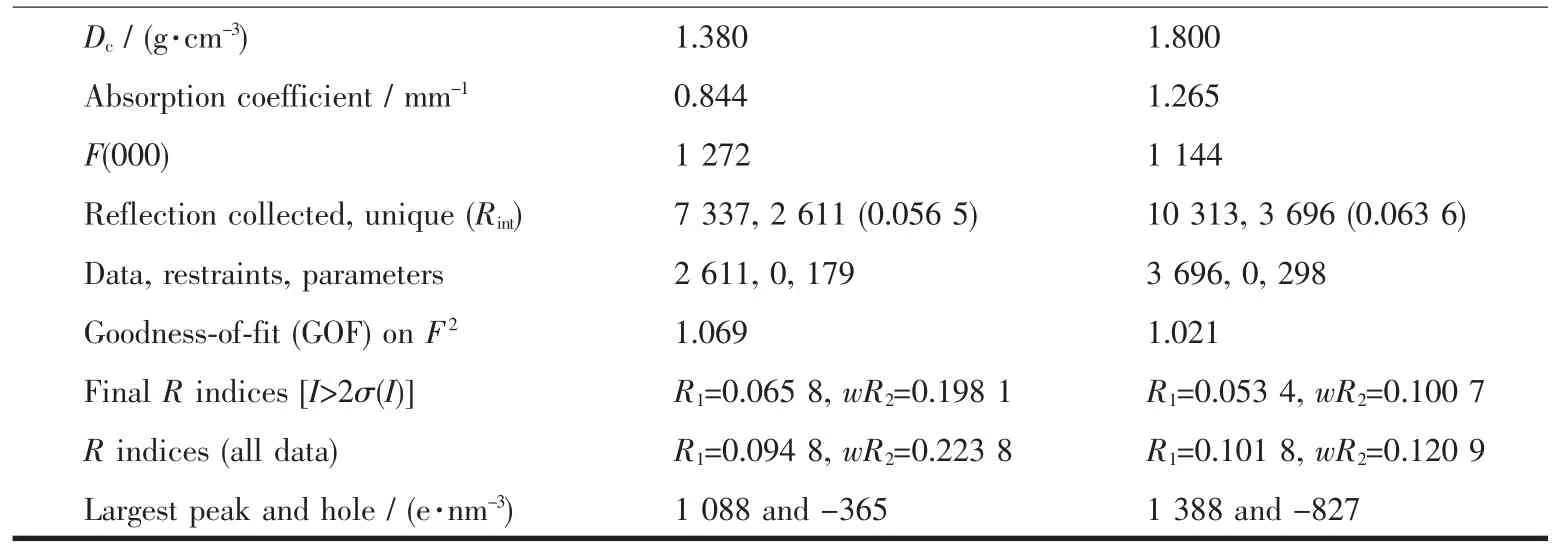
Continued Table 1
2 Results and discussion
2.1Crystal structures description
The ORTEP drawing of complexes 1 and 2 is shown in Fig.1. Selected bond distances and angles are listed in Table 2. Hydrogen bonds information is in Table 3. The lengths of C-S bond are in the range of 0.166 7(7)~0.171 5(6) nm in complexes 1 and 2, showing that the ligand HL has thiolated and deprotonated in both complexes[14].
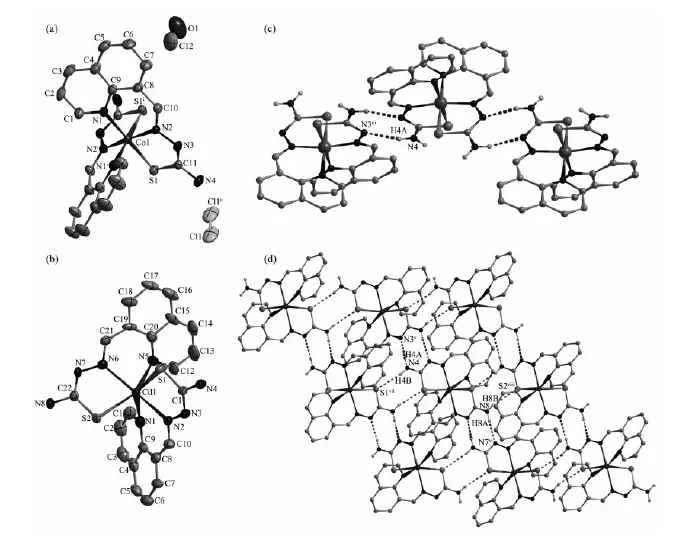
Fig.1 ORTEP drawing of 1 (a) and 2 (b) with 30% thermal ellipsoids; (c) Chain-like structure along c axis formed via pairs of N-H…N hydrogen bonds in complex 1; (d) Extended 2D supermolecular structure in complex 2
As shown in Fig.1a, the asymmetric unit of complex 1 contains a half of the molecule with theCoion situated on a two-fold rotational axis. The centre Coion is coordinated by two anionic TSC ligands with N2S donor sets, and thus possesses a distorted octahedron coordination geometry. There exists one free chloride anion(occupying two positions) in the outside of the complex for charge balance, although complex 1 is prepared by the reaction of the TSC ligand HL with CoCl2in methanol medium. This is a normal phenomenon in the Cocomplexes with TSC ligands reported in literature[14]. The distances of Co-N/S bonds were in the range of 0.188 0(6)~0.220 4(4) nm, which were comparable with those found in the reported complexes with similar donor set. In the crystal, pairs of intermolecular N -H…N hydrogen bonds link the complex cations into one-dimentional chains along c axis (Fig. 1c). Furthermore, a 3D supermolecular network is formed via the N-H…Cl hydrogen bonds between the chains and free chlorides. The intermolecular O-H…Cl hydrogen bonds between the crystal methanol molecules and chloride anions are also present (Table 3).
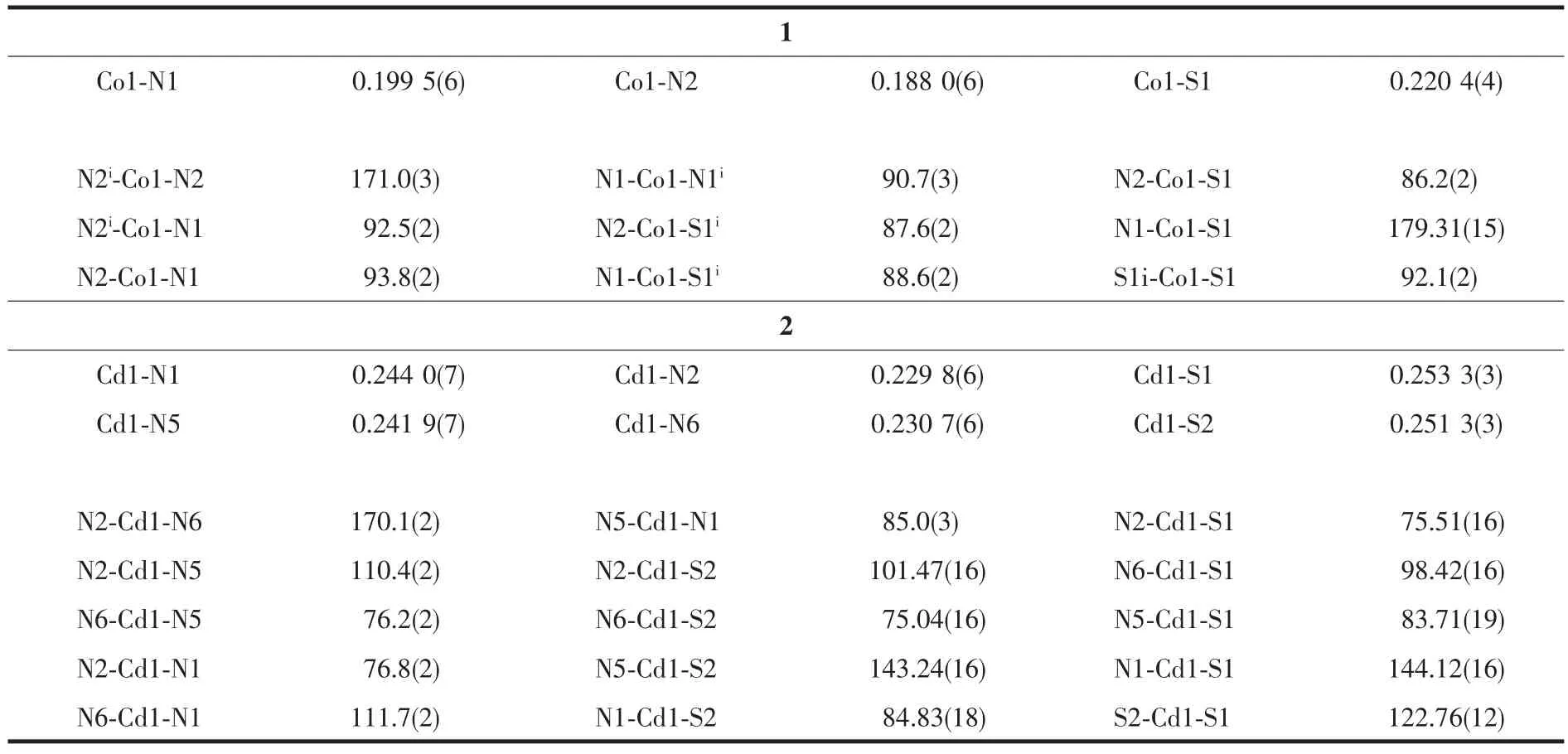
Table 2 Selected bond lengths (nm) and angles (°) in complexes 1 and 2
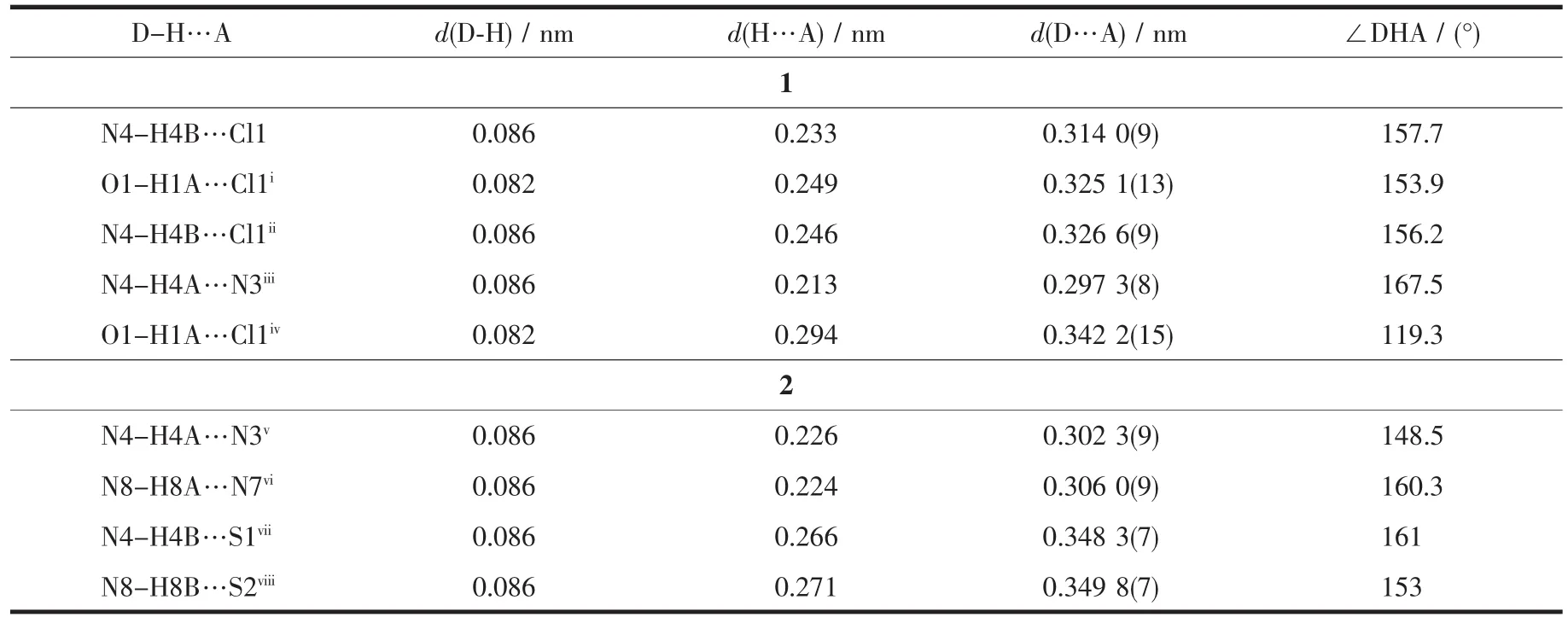
Table 3 Hydrogen bonds information in complexes 1 and 2
The coordination environment of central Cdion in complex 2 (Fig.1b) is quite similar as that ofCoion in complex 1. The distances of Cd-N/S bonds were in the normal range (0.229 8(6)~0.253 3(3) nm). In the crystal of 2, the intermolecular N-H…N hydrogen bonds link the complexes into chains, which were further linked with each other via intermolecular N-H…S hydrogen bonds to form extended 2D supermolecular network, as illustrated in Fig.1d.
2.2IR spectra
The infrared spectral bands most useful for determining the mode of coordination of the ligands are the ν(N=C),ν(N=C, pyrazine) and ν(S=C) vibrations. Such three bonds of the free TSC ligand is found at 1 594, 1 532 and 816 cm-1, while they shifts to lower frequency in complexes 1 and 2, clearly indicating the coordination of imine nitrogen, quinoline nitrogen and sulfur atoms[14]. It is in accordance with the X-ray diffraction analysis result.
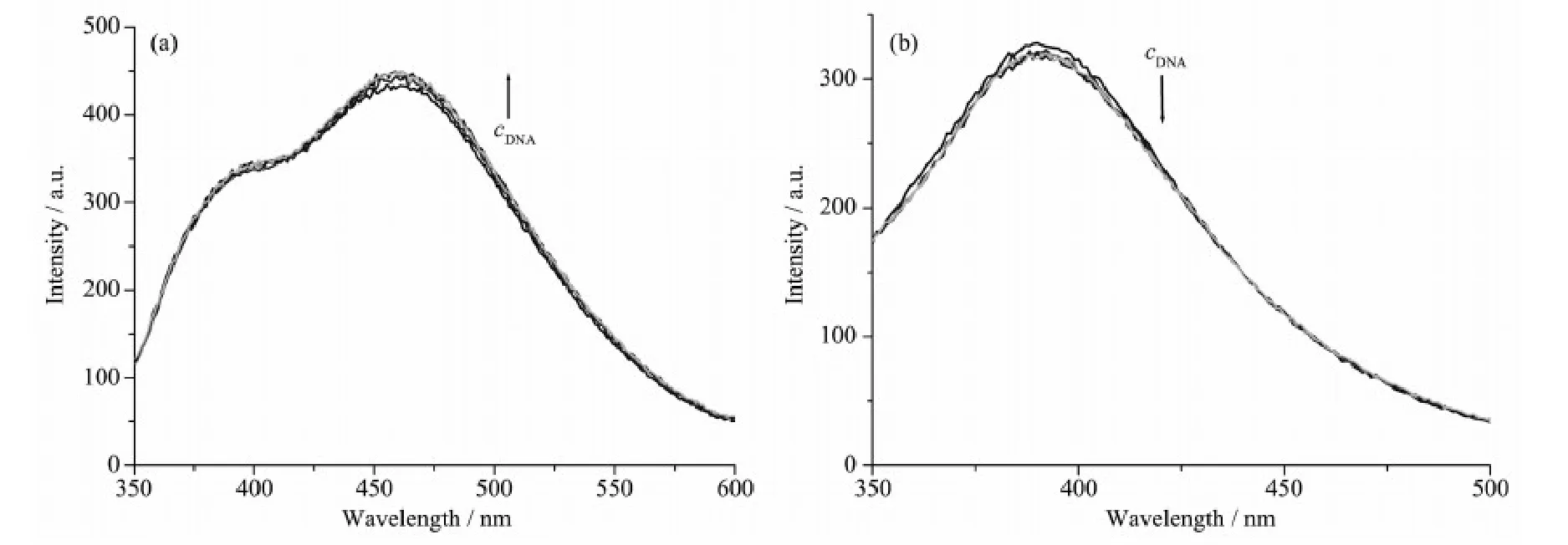
Fig.3 Emission enhancement spectra of the complexes 1 (a) and 2 (b) in Tris-HCl buffer (pH=7.2) upon addition of CT-DNA
2.3DNA-binding studies
The application of electronic absorption spectroscopy is one of the most useful techniques in DNA-binding studies. The UV absorption spectra would change in accordance with the environmental condition changes, since the stacking interactions happened between the complexes and DNA[14,17-18]. As shown in Fig.2, with increasing DNA concentration, hypochromic (1) or hyperchromic (2) effect was observed, which indicated molecular level interactions between the complexes and DNA. In addition, the lack of the red shift suggests that the major binding modes of both complexes with DNA are not classical intercalative interaction[18]. The intrinsic binding constant (Kb) of 1 was determined to be 8.81×103L·mol-1according to the literature method[14,17], which is comparable with some Cocomplexes with TSCs ligands[14]. However,the absorption intensities change of 2 is obscure, thus its binding constant cannot be obtained reasonably.
The emission experiment has been widely used to further confirm the interaction between the complexes and CT-DNA. The results of fluorescence titration spectra have also been confirmed to be effective for characterizing the binding mode of the metal complexes to DNA[17-18]. Fig.3 shows the results of the emission titration curve of the complexes with CTDNA. An increase in DNA concentration resulted slight increase and decrease in the emission intensity of complexes 1 and 2, respectively. Combining with the electronic absorption spectroscopy titration results and according to the literature, it can be roughly concluded that complexes 1 and 2 bind to DNA via electrostatic intercalation[17]and partial intercalation[18]modes, respectively.
3 Conclusions
In summary, two transition metal complexes [CoL2]Cl·2CH3OH (1) and [CdL2] (2) were synthesized and characterized. The metal ion in each complex with a distorted octahedron geometry is surrounded by two anionic thiosemicarbazone ligand with N2S donor set. Complexes 1 and 2 can bind to DNA via electrostatic intercalation and partial intercalation modes, respectively. This indicates that both complexes have potential pharmaceutical activity.
References:
[1] Lobana T S, Sharma R, Bawa G, et al. Coord. Chem. Rev., 2009,253:977-1055
[2] Soares M A, Lessa J A, Mendes I C, et al. Bioorg. Med. Chem., 2012,20:3396-3409
[3] Huang H, Chen Q, Ku X, et al. J. Med. Chem., 2010,53: 3048-3064
[4] Qi J, Su L, Gou Y, et al. Eur. J. Med. Chem., 2015,96:360-368
[5] Zeglis B M, Divilov V, Lewis J S. J. Med. Chem., 2011,54: 2391-2398
[6] Li M X, Zhang L Z, Yang M, et al. Bioorg. Med. Chem. Lett., 2012,22:2418-2423
[7] Li M X, Zhang L Z, Zhang D, et al. Eur. J. Med. Chem., 2011,46:4383-4390
[8] Li M X, Chen C L, Zhang D, et al. Eur. J. Med. Chem., 2010,45:3169-3177
[9] Kalinowski D S, Yu Y, Sharpe P C, et al. J. Med. Chem., 2007,50:3716-3729
[10]Lovejoy D B, Sharp D M, Seebacher N, et al. J. Med. Chem., 2012,55:7230-7244
[11]MIN Rui(閔睿), FAN Xiao-Rui(范曉瑞), ZHOU Pan(周攀), et al. Chinese J. Inorg. Chem.(無機(jī)化學(xué)學(xué)報(bào)), 2014,30: 1171-1177
[12]Bourosh P N, Revenko M D, Stratulat E F, et al. Russ. J. Inorg. Chem., 2014,59:545-557
[13]Revenko M D, Bourosh P N, Stratulat E F, et al. Russ. J. Inorg. Chem., 2010,55:1387-1397
[14]Ramachandran E, Thomas S P, Poornima P, et al. Eur. J. Med. Chem., 2012,50:405-415
[15]Sheldrick G M. SADABS, University of G?ttingen, Germany, 1996.
[16]Sheldrick G M. SHELX-97, Program for the Solution and the Refinement of Crystal Structures, University of G?ttingen, Germany, 1997.
[17]CAO Feng-Pu(曹豐璞), DING Cheng-Hua(丁呈華), LIU Wen -Min(柳文敏), et al. Chinese J. Inorg. Chem.(無機(jī)化學(xué)學(xué)報(bào)), 2011,27:343-347
[18]Wang Y, Yang Z Y, Chen Z N. Bioorg. Med. Chem. Lett., 2008,18:298-303
毛盼東1閆玲玲*,1王文靜2楊倩倩2崔猛強(qiáng)1王元*,1吳偉娜1
(1河南理工大學(xué)物理化學(xué)學(xué)院,焦作454000)
(2河南理工大學(xué)材料科學(xué)與工程學(xué)院,焦作454000)
合成并通過單晶衍射、元素分析、紅外光譜表征了配合物[CoL2]Cl·2CH3OH (1)和[CdL2] (2)的結(jié)構(gòu)(HL為喹啉-8-甲醛縮硫代氨基脲)。單晶衍射結(jié)果表明,配合物1和2中金屬離子采取相同的配位模式,分別與來自硫醇化脫質(zhì)子的配體L-的4個(gè)N原子和2個(gè)S原子配位,采取扭曲的八面體配位構(gòu)型。配合物1和2能夠與DNA結(jié)合,結(jié)合模式分別為靜電結(jié)合和部分插入。
喹啉;縮氨基硫脲;配合物;晶體結(jié)構(gòu);DNA結(jié)合性質(zhì)
O614.81+2;O614.24+2
A
1001-4861(2016)03-0555-06
10.11862/CJIC.2016.064
2015-11-18。收修改稿日期:2016-01-15。
國(guó)家自然科學(xué)基金(No.21001040)、河南省教育廳自然科學(xué)基金(No.12B150011,14B150029)資助項(xiàng)目。
*通信聯(lián)系人。E-mail:yll@hpu.edu.cn; wangyuan08@hpu.edu.cn;會(huì)員登記號(hào):S06N4036M1112。
 無機(jī)化學(xué)學(xué)報(bào)2016年3期
無機(jī)化學(xué)學(xué)報(bào)2016年3期
- 無機(jī)化學(xué)學(xué)報(bào)的其它文章
- Photocatalytic Killing Effect of Fe3O4-TiO2Nanoparticles on Hepatoma Carcinoma Cells for Targeting Photodynamic Therapy
- Four- and Three-Fold Interpenetrated (3,4)-Connected d10Metal Coordination Polymers Constructed by 3,5-Bis(pyridin-4-ylmethoxy)benzoic Acid and Aromatic Dicarboxylic Acid Ligands
- Polypyridyl Ligands-Based Double-Decker Triggered by Silver(Ⅰ)Coordination: Crystal Structures and Spectroscopic Analysis
- Porous Tremella-like NiO on Conductive Substrates with High Electrochemical Performance
- Promotional Effect of Zr on Thermal Stability of CeTiOxMonolith Catalyst for Selective Catalytic Reduction of NOxwith Ammonia
- A π-Conjugated Organic Molecule: BDOBC16 Assembly in Silylated SBA-15 and Luminescent Properties
